| 1 |
|
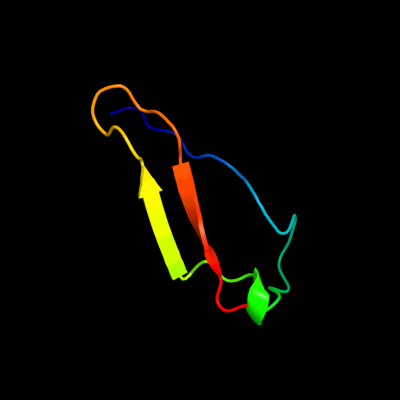
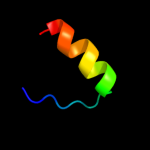
PDB 3dw8 chain B
Region: 35 - 75
Aligned: 39
Modelled: 41
Confidence: 22.0%
Identity: 15%
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:serine/threonine-protein phosphatase 2a 55 kda regulatory
PDBTitle: structure of a protein phosphatase 2a holoenzyme with b55 subunit
Phyre2
| 2 |
|
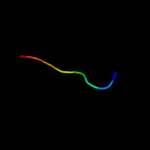
PDB 1jl3 chain A
Region: 25 - 32
Aligned: 8
Modelled: 8
Confidence: 18.7%
Identity: 38%
Fold: Phosphotyrosine protein phosphatases I-like
Superfamily: Phosphotyrosine protein phosphatases I
Family: Low-molecular-weight phosphotyrosine protein phosphatases
Phyre2
| 3 |
|
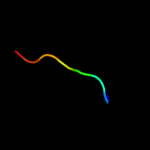
PDB 3nct chain C
Region: 10 - 32
Aligned: 19
Modelled: 23
Confidence: 17.3%
Identity: 32%
PDB header:dna binding protein, chaperone
Chain: C: PDB Molecule:protein psib;
PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
Phyre2
| 4 |
|
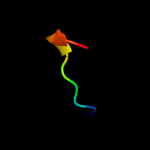
PDB 2krc chain A
Region: 58 - 77
Aligned: 17
Modelled: 20
Confidence: 16.2%
Identity: 35%
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit delta;
PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
Phyre2
| 5 |
|
PDB 2noc chain A domain 1
Region: 72 - 83
Aligned: 12
Modelled: 12
Confidence: 14.8%
Identity: 25%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 6 |
|
PDB 2pxg chain A
Region: 37 - 67
Aligned: 31
Modelled: 31
Confidence: 9.2%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane protein;
PDBTitle: nmr solution structure of omla
Phyre2
| 7 |
|
PDB 2jv5 chain A
Region: 43 - 51
Aligned: 9
Modelled: 9
Confidence: 8.5%
Identity: 33%
PDB header:protein binding
Chain: A: PDB Molecule:reticulon-4;
PDBTitle: nogo54
Phyre2
| 8 |
|
PDB 2kxx chain A
Region: 36 - 69
Aligned: 33
Modelled: 34
Confidence: 6.7%
Identity: 24%
PDB header:protein binding
Chain: A: PDB Molecule:small protein a;
PDBTitle: nmr structure of escherichia coli bame, a lipoprotein component of the2 beta-barrel assembly machinery complex
Phyre2
| 9 |
|
PDB 2igi chain A domain 1
Region: 53 - 67
Aligned: 15
Modelled: 15
Confidence: 6.6%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Ribonuclease H-like
Family: DnaQ-like 3'-5' exonuclease
Phyre2
| 10 |
|
PDB 1y8a chain A
Region: 38 - 57
Aligned: 20
Modelled: 20
Confidence: 6.5%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af1437;
PDBTitle: structure of gene product af1437 from archaeoglobus fulgidus
Phyre2
| 11 |
|
PDB 2ko2 chain A
Region: 43 - 51
Aligned: 9
Modelled: 9
Confidence: 6.4%
Identity: 33%
PDB header:membrane protein
Chain: A: PDB Molecule:reticulon-4;
PDBTitle: nogo66
Phyre2
| 12 |
|
PDB 1x9d chain A
Region: 39 - 50
Aligned: 11
Modelled: 12
Confidence: 6.4%
Identity: 55%
PDB header:hydrolase
Chain: A: PDB Molecule:endoplasmic reticulum mannosyl-oligosaccharide 1,
PDBTitle: crystal structure of human class i alpha-1,2-mannosidase in2 complex with thio-disaccharide substrate analogue
Phyre2
| 13 |
|
PDB 1x9d chain A domain 1
Region: 39 - 50
Aligned: 11
Modelled: 12
Confidence: 6.4%
Identity: 55%
Fold: alpha/alpha toroid
Superfamily: Seven-hairpin glycosidases
Family: Class I alpha-1;2-mannosidase, catalytic domain
Phyre2
| 14 |
|
PDB 2ker chain A
Region: 37 - 44
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 50%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:alpha-amylase inhibitor z-2685;
PDBTitle: alpha-amylase inhibitor parvulustat (z-2685) from2 streptomyces parvulus
Phyre2
| 15 |
|
PDB 1ok0 chain A
Region: 37 - 44
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 50%
Fold: alpha-Amylase inhibitor tendamistat
Superfamily: alpha-Amylase inhibitor tendamistat
Family: alpha-Amylase inhibitor tendamistat
Phyre2
| 16 |
|
PDB 3asi chain A
Region: 74 - 81
Aligned: 8
Modelled: 8
Confidence: 5.8%
Identity: 63%
PDB header:cell adhesion
Chain: A: PDB Molecule:neurexin-1-alpha;
PDBTitle: alpha-neurexin-1 ectodomain fragment; lns5-egf3-lns6
Phyre2
| 17 |
|
PDB 1hcu chain A
Region: 38 - 50
Aligned: 12
Modelled: 13
Confidence: 5.5%
Identity: 33%
Fold: alpha/alpha toroid
Superfamily: Seven-hairpin glycosidases
Family: Class I alpha-1;2-mannosidase, catalytic domain
Phyre2
| 18 |
|
PDB 3d3r chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 55%
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
Phyre2
| 19 |
|
PDB 1p2z chain A domain 2
Region: 30 - 44
Aligned: 15
Modelled: 15
Confidence: 5.2%
Identity: 53%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Group II dsDNA viruses VP
Family: Adenovirus hexon
Phyre2
| 20 |
|
PDB 1hnf chain A domain 2
Region: 52 - 60
Aligned: 9
Modelled: 9
Confidence: 5.2%
Identity: 78%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: C2 set domains
Phyre2
| 21 |
|
| 22 |
|