| 1 |
|
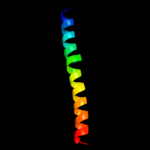
PDB 3bt6 chain B
Region: 1 - 40
Aligned: 40
Modelled: 40
Confidence: 17.8%
Identity: 25%
PDB header:viral protein
Chain: B: PDB Molecule:influenza b hemagglutinin (ha);
PDBTitle: crystal structure of influenza b virus hemagglutinin
Phyre2
| 2 |
|
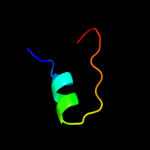
PDB 1htm chain B
Region: 1 - 40
Aligned: 40
Modelled: 40
Confidence: 16.1%
Identity: 28%
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin ha2 chain;
PDBTitle: structure of influenza haemagglutinin at the ph of membrane2 fusion
Phyre2
| 3 |
|
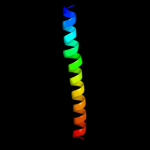
PDB 1ru7 chain B
Region: 1 - 40
Aligned: 40
Modelled: 40
Confidence: 15.2%
Identity: 20%
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin;
PDBTitle: 1934 human h1 hemagglutinin
Phyre2
| 4 |
|
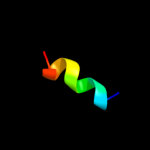
PDB 2wr2 chain B
Region: 1 - 40
Aligned: 40
Modelled: 40
Confidence: 14.7%
Identity: 23%
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin;
PDBTitle: structure of influenza h2 avian hemagglutinin with avian2 receptor
Phyre2
| 5 |
|
PDB 1jsd chain B
Region: 1 - 40
Aligned: 40
Modelled: 40
Confidence: 12.1%
Identity: 23%
PDB header:viral protein
Chain: B: PDB Molecule:haemagglutinin (ha2 chain);
PDBTitle: crystal structure of swine h9 haemagglutinin
Phyre2
| 6 |
|
PDB 3m5g chain D
Region: 2 - 40
Aligned: 38
Modelled: 39
Confidence: 10.9%
Identity: 24%
PDB header:viral protein
Chain: D: PDB Molecule:hemagglutinin;
PDBTitle: crystal structure of a h7 influenza virus hemagglutinin
Phyre2
| 7 |
|
PDB 2wrh chain I
Region: 2 - 40
Aligned: 38
Modelled: 39
Confidence: 10.2%
Identity: 24%
PDB header:viral protein
Chain: I: PDB Molecule:hemagglutinin ha2 chain;
PDBTitle: structure of h1 duck albert hemagglutinin with human2 receptor
Phyre2
| 8 |
|
PDB 1t6u chain A
Region: 13 - 39
Aligned: 27
Modelled: 27
Confidence: 8.5%
Identity: 22%
Fold: Four-helical up-and-down bundle
Superfamily: Nickel-containing superoxide dismutase, NiSOD
Family: Nickel-containing superoxide dismutase, NiSOD
Phyre2
| 9 |
|
PDB 1mql chain B
Region: 2 - 40
Aligned: 38
Modelled: 39
Confidence: 8.5%
Identity: 18%
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin ha2 chain;
PDBTitle: bha of ukr/63
Phyre2
| 10 |
|
PDB 1reg chain X
Region: 20 - 40
Aligned: 21
Modelled: 21
Confidence: 6.5%
Identity: 33%
Fold: Ferredoxin-like
Superfamily: Translational regulator protein regA
Family: Translational regulator protein regA
Phyre2
| 11 |
|
PDB 1hge chain D
Region: 2 - 40
Aligned: 38
Modelled: 39
Confidence: 6.5%
Identity: 18%
PDB header:viral protein
Chain: D: PDB Molecule:hemagglutinin, (g135r), ha1 chain;
PDBTitle: binding of influenza virus hemagglutinin to analogs of its cell-2 surface receptor, sialic acid: analysis by proton nuclear magnetic3 resonance spectroscopy and x-ray crystallography
Phyre2
| 12 |
|
PDB 2rmi chain A
Region: 60 - 70
Aligned: 11
Modelled: 11
Confidence: 6.3%
Identity: 64%
PDB header:neuropeptide
Chain: A: PDB Molecule:astressin;
PDBTitle: 3d nmr structure of astressin
Phyre2
| 13 |
|
PDB 1ha0 chain A
Region: 2 - 40
Aligned: 38
Modelled: 39
Confidence: 6.3%
Identity: 18%
PDB header:viral protein
Chain: A: PDB Molecule:protein (hemagglutinin precursor);
PDBTitle: hemagglutinin precursor ha0
Phyre2