| 1 |
|
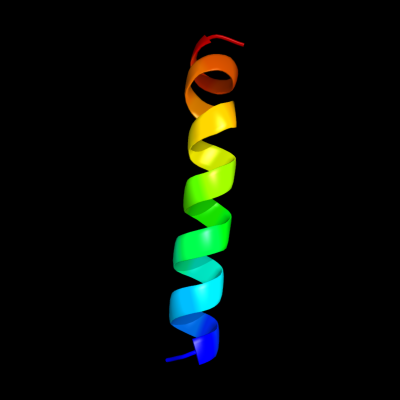
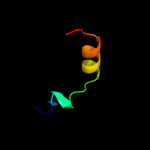
PDB 2jp3 chain A
Region: 41 - 64
Aligned: 24
Modelled: 24
Confidence: 12.1%
Identity: 29%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 2 |
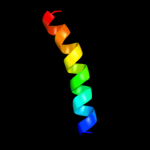
|
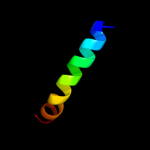
PDB 1tjn chain A
Region: 49 - 74
Aligned: 26
Modelled: 26
Confidence: 11.2%
Identity: 27%
PDB header:lyase
Chain: A: PDB Molecule:sirohydrochlorin cobaltochelatase;
PDBTitle: crystal structure of hypothetical protein af0721 from archaeoglobus2 fulgidus
Phyre2
| 3 |
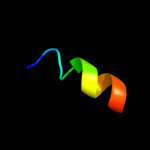
|
PDB 1tjn chain A
Region: 49 - 74
Aligned: 26
Modelled: 26
Confidence: 11.2%
Identity: 27%
Fold: Chelatase-like
Superfamily: Chelatase
Family: CbiX-like
Phyre2
| 4 |
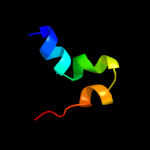
|
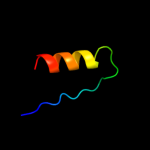
PDB 1i0v chain A
Region: 15 - 29
Aligned: 15
Modelled: 15
Confidence: 9.5%
Identity: 33%
Fold: Microbial ribonucleases
Superfamily: Microbial ribonucleases
Family: Fungal ribonucleases
Phyre2
| 5 |
|
PDB 2drp chain A domain 2
Region: 25 - 30
Aligned: 6
Modelled: 6
Confidence: 8.9%
Identity: 83%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 6 |
|
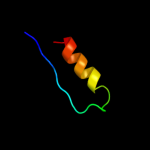
PDB 3a9l chain B
Region: 16 - 39
Aligned: 24
Modelled: 24
Confidence: 8.3%
Identity: 21%
PDB header:hydrolase
Chain: B: PDB Molecule:poly-gamma-glutamate hydrolase;
PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
Phyre2
| 7 |
|
PDB 2crg chain A domain 1
Region: 44 - 68
Aligned: 23
Modelled: 25
Confidence: 7.5%
Identity: 26%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Myb/SANT domain
Phyre2
| 8 |
|
PDB 2jo1 chain A
Region: 41 - 62
Aligned: 22
Modelled: 22
Confidence: 7.3%
Identity: 23%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 9 |
|
PDB 2kaa chain A
Region: 18 - 29
Aligned: 12
Modelled: 12
Confidence: 6.2%
Identity: 33%
PDB header:toxin
Chain: A: PDB Molecule:hirsutellin a;
PDBTitle: solution structure of hirsutellin a from hirsutella2 thompsonii
Phyre2
| 10 |
|
PDB 2oqb chain A
Region: 26 - 34
Aligned: 9
Modelled: 9
Confidence: 6.2%
Identity: 56%
PDB header:transferase,gene regulation
Chain: A: PDB Molecule:histone-arginine methyltransferase carm1;
PDBTitle: crystal structure of the n-terminal domain of coactivator-associated2 methyltransferase 1 (carm1)
Phyre2
| 11 |
|
PDB 1isu chain A
Region: 78 - 87
Aligned: 10
Modelled: 10
Confidence: 5.9%
Identity: 30%
Fold: HIPIP (high potential iron protein)
Superfamily: HIPIP (high potential iron protein)
Family: HIPIP (high potential iron protein)
Phyre2
| 12 |
|
PDB 3kdp chain G
Region: 42 - 54
Aligned: 13
Modelled: 13
Confidence: 5.7%
Identity: 46%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 13 |
|
PDB 3kdp chain H
Region: 42 - 54
Aligned: 13
Modelled: 13
Confidence: 5.7%
Identity: 46%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 14 |
|
PDB 3m8e chain A
Region: 60 - 86
Aligned: 20
Modelled: 27
Confidence: 5.7%
Identity: 50%
PDB header:dna binding protein
Chain: A: PDB Molecule:putative dna-binding protein;
PDBTitle: protein structure of type iii plasmid segregation tubr
Phyre2