| 1 |
|
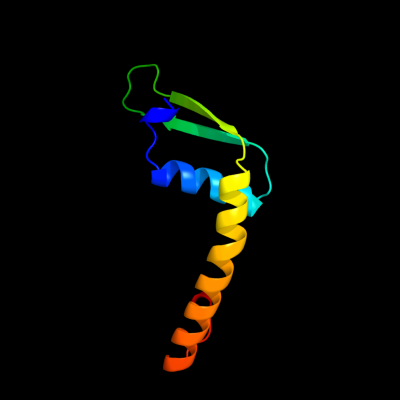
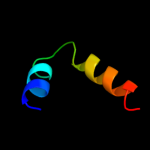
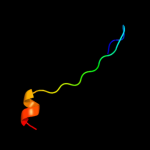
PDB 1avq chain A
Region: 7 - 87
Aligned: 81
Modelled: 81
Confidence: 100.0%
Identity: 96%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: lambda exonuclease
Phyre2
| 2 |
|
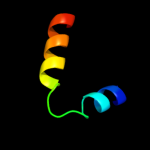
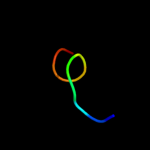
PDB 3k93 chain A
Region: 15 - 79
Aligned: 65
Modelled: 65
Confidence: 97.4%
Identity: 20%
PDB header:hydrolase
Chain: A: PDB Molecule:phage related exonuclease;
PDBTitle: crystal structure of phage related exonuclease (yp_719632.1) from2 haemophilus somnus 129pt at 2.15 a resolution
Phyre2
| 3 |
|
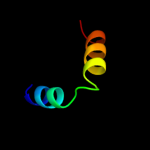
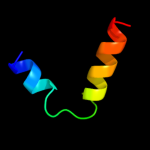
PDB 1jbj chain A domain 2
Region: 29 - 49
Aligned: 21
Modelled: 21
Confidence: 15.8%
Identity: 24%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 4 |
|
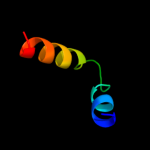
PDB 2gts chain A domain 1
Region: 50 - 82
Aligned: 33
Modelled: 33
Confidence: 14.0%
Identity: 15%
Fold: Ferritin-like
Superfamily: HP0062-like
Family: HP0062-like
Phyre2
| 5 |
|
PDB 1zaw chain U
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.5%
Identity: 15%
PDB header:structural protein
Chain: U: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
Phyre2
| 6 |
|
PDB 1zaw chain W
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.5%
Identity: 15%
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
Phyre2
| 7 |
|
PDB 1zav chain U domain 1
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.5%
Identity: 15%
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Phyre2
| 8 |
|
PDB 1zax chain U
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.5%
Identity: 15%
PDB header:structural protein
Chain: U: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
Phyre2
| 9 |
|
PDB 1zav chain U
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.5%
Identity: 15%
PDB header:structural protein
Chain: U: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
Phyre2
| 10 |
|
PDB 1zax chain W
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.2%
Identity: 15%
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
Phyre2
| 11 |
|
PDB 1zav chain V
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.0%
Identity: 15%
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
Phyre2
| 12 |
|
PDB 1zax chain V
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 10.0%
Identity: 15%
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
Phyre2
| 13 |
|
PDB 1zav chain W
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 9.7%
Identity: 15%
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
Phyre2
| 14 |
|
PDB 1zaw chain V
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 8.7%
Identity: 15%
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
Phyre2
| 15 |
|
PDB 1dd3 chain D
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 8.6%
Identity: 15%
PDB header:ribosome
Chain: D: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
Phyre2
| 16 |
|
PDB 1dd3 chain C
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 8.6%
Identity: 15%
PDB header:ribosome
Chain: C: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
Phyre2
| 17 |
|
PDB 2ktl chain A
Region: 39 - 59
Aligned: 21
Modelled: 21
Confidence: 8.2%
Identity: 19%
PDB header:ligase
Chain: A: PDB Molecule:tyrosyl-trna synthetase;
PDBTitle: structure of c-terminal domain from mttyrrs of a. nidulans
Phyre2
| 18 |
|
PDB 1qzf chain A domain 2
Region: 78 - 87
Aligned: 10
Modelled: 10
Confidence: 7.7%
Identity: 50%
Fold: Thymidylate synthase/dCMP hydroxymethylase
Superfamily: Thymidylate synthase/dCMP hydroxymethylase
Family: Thymidylate synthase/dCMP hydroxymethylase
Phyre2
| 19 |
|
PDB 1dd4 chain C
Region: 57 - 83
Aligned: 27
Modelled: 27
Confidence: 6.4%
Identity: 15%
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Phyre2
| 20 |
|
PDB 1j3k chain C
Region: 78 - 87
Aligned: 10
Modelled: 10
Confidence: 5.6%
Identity: 50%
Fold: Thymidylate synthase/dCMP hydroxymethylase
Superfamily: Thymidylate synthase/dCMP hydroxymethylase
Family: Thymidylate synthase/dCMP hydroxymethylase
Phyre2
| 21 |
|