| 1 |
|
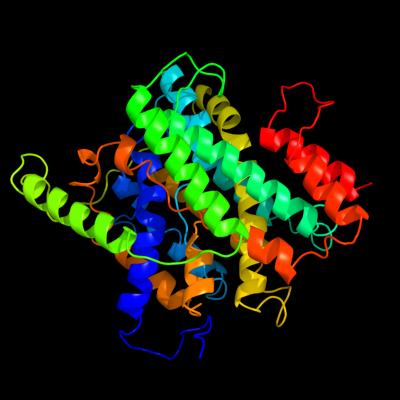
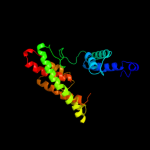
PDB 3qe7 chain A
Region: 17 - 417
Aligned: 378
Modelled: 395
Confidence: 100.0%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:uracil permease;
PDBTitle: crystal structure of uracil transporter--uraa
Phyre2
| 2 |
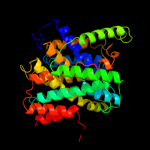
|
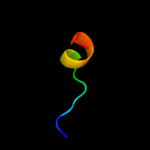
PDB 3lpz chain A
Region: 299 - 312
Aligned: 14
Modelled: 14
Confidence: 12.7%
Identity: 21%
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
Phyre2
| 3 |
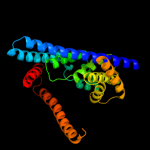
|
PDB 3org chain B
Region: 135 - 403
Aligned: 254
Modelled: 254
Confidence: 12.6%
Identity: 10%
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
Phyre2
| 4 |
|
PDB 2ht2 chain B
Region: 13 - 231
Aligned: 206
Modelled: 219
Confidence: 9.5%
Identity: 12%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 5 |
|
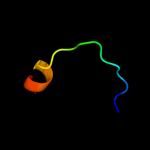
PDB 1hlv chain A domain 2
Region: 6 - 18
Aligned: 13
Modelled: 13
Confidence: 8.2%
Identity: 31%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Centromere-binding
Phyre2
| 6 |
|
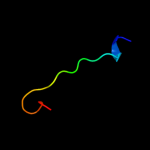
PDB 1iuf chain A domain 2
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 8.1%
Identity: 18%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Centromere-binding
Phyre2
| 7 |
|
PDB 1w3g chain A
Region: 14 - 30
Aligned: 17
Modelled: 17
Confidence: 7.2%
Identity: 18%
PDB header:toxin/lectin
Chain: A: PDB Molecule:hemolytic lectin from laetiporus sulphureus;
PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
Phyre2
| 8 |
|
PDB 3iys chain A
Region: 15 - 30
Aligned: 16
Modelled: 16
Confidence: 7.0%
Identity: 25%
PDB header:virus
Chain: A: PDB Molecule:major capsid protein vp1;
PDBTitle: homology model of avian polyomavirus asymmetric unit
Phyre2