| 1 |
|
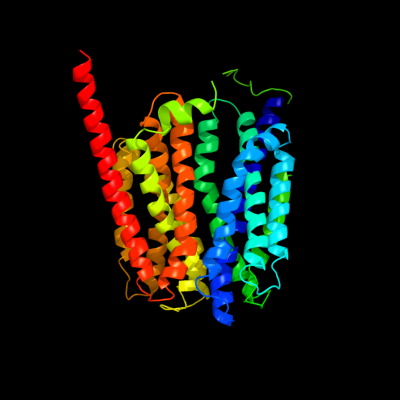
PDB 1pw4 chain A
Region: 2 - 403
Aligned: 401
Modelled: 402
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
PDB 2gfp chain A
Region: 15 - 384
Aligned: 370
Modelled: 370
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 3 |
|
PDB 3o7p chain A
Region: 9 - 389
Aligned: 373
Modelled: 381
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
PDB 1pv7 chain A
Region: 13 - 406
Aligned: 393
Modelled: 394
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 16 - 388
Aligned: 373
Modelled: 373
Confidence: 99.9%
Identity: 15%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 245 - 420
Aligned: 174
Modelled: 176
Confidence: 65.2%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 3rko chain N
Region: 7 - 415
Aligned: 396
Modelled: 396
Confidence: 54.5%
Identity: 9%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 8 |
|
PDB 3qnq chain D
Region: 356 - 407
Aligned: 52
Modelled: 52
Confidence: 29.0%
Identity: 21%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 9 |
|
PDB 2g9p chain A
Region: 67 - 80
Aligned: 14
Modelled: 14
Confidence: 16.7%
Identity: 50%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 10 |
|
PDB 1zxa chain B
Region: 394 - 419
Aligned: 26
Modelled: 26
Confidence: 8.9%
Identity: 12%
PDB header:transferase
Chain: B: PDB Molecule:cgmp-dependent protein kinase 1, alpha isozyme;
PDBTitle: solution structure of the coiled-coil domain of cgmp-2 dependent protein kinase ia
Phyre2
| 11 |
|
PDB 3hd6 chain A
Region: 198 - 418
Aligned: 217
Modelled: 221
Confidence: 8.0%
Identity: 14%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 12 |
|
PDB 1odr chain A
Region: 404 - 421
Aligned: 18
Modelled: 18
Confidence: 7.7%
Identity: 22%
PDB header:lipid transport
Chain: A: PDB Molecule:apoa-i peptide;
PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:dpc mole ratio3 of 1:40
Phyre2
| 13 |
|
PDB 1odq chain A
Region: 404 - 421
Aligned: 18
Modelled: 18
Confidence: 7.7%
Identity: 22%
PDB header:lipid transport
Chain: A: PDB Molecule:apoa-i peptide;
PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 3.7, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
Phyre2
| 14 |
|
PDB 1odp chain A
Region: 404 - 421
Aligned: 18
Modelled: 18
Confidence: 7.7%
Identity: 22%
PDB header:lipid transport
Chain: A: PDB Molecule:apoa-i peptide;
PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.6, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
Phyre2
| 15 |
|
PDB 1vkz chain A domain 1
Region: 399 - 421
Aligned: 23
Modelled: 23
Confidence: 6.0%
Identity: 13%
Fold: Barrel-sandwich hybrid
Superfamily: Rudiment single hybrid motif
Family: BC C-terminal domain-like
Phyre2
| 16 |
|
PDB 1rh1 chain A domain 2
Region: 9 - 77
Aligned: 67
Modelled: 69
Confidence: 5.5%
Identity: 13%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 17 |
|
PDB 2knc chain B
Region: 369 - 413
Aligned: 45
Modelled: 45
Confidence: 5.3%
Identity: 13%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2