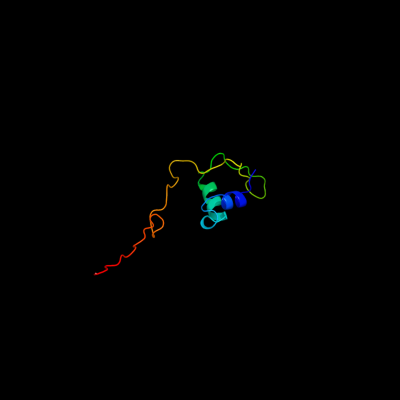
1 d1pm6a_
100.0
38
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like2 d1rh6a_
99.9
39
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like3 c1z4hA_
90.8
15
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: tor inhibition protein;PDBTitle: the response regulator tori belongs to a new family of2 atypical excisionase
4 d1r8ea1
88.6
21
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators5 c2zhhA_
86.2
23
PDB header: transcriptionChain: A: PDB Molecule: redox-sensitive transcriptional activator soxr;PDBTitle: crystal structure of soxr
6 c3hh0C_
83.9
30
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator, merr family;PDBTitle: crystal strucure of a transcriptional regulator, merr family2 from bacillus cereus
7 c3d6zA_
81.9
25
PDB header: transcription regulator/dnaChain: A: PDB Molecule: multidrug-efflux transporter 1 regulator;PDBTitle: crystal structure of r275e mutant of bmrr bound to dna and rhodamine
8 d1q06a_
70.1
14
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators9 c3gpvA_
68.2
26
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, merr family;PDBTitle: crystal structure of a transcriptional regulator, merr2 family from bacillus thuringiensis
10 c1y6uA_
68.0
25
PDB header: dna binding proteinChain: A: PDB Molecule: excisionase from transposon tn916;PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
11 c3qaoA_
67.5
27
PDB header: transcription regulatorChain: A: PDB Molecule: merr-like transcriptional regulator;PDBTitle: the crystal structure of the n-terminal domain of a merr-like2 transcriptional regulator from listeria monocytogenes egd-e
12 c2vz4A_
65.0
19
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional activator tipa;PDBTitle: the n-terminal domain of merr-like protein tipal bound to2 promoter dna
13 d1r8da_
61.6
18
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators14 c2jmlA_
56.9
24
PDB header: transcriptionChain: A: PDB Molecule: dna binding domain/transcriptional regulator;PDBTitle: solution structure of the n-terminal domain of cara repressor
15 c3gp4B_
50.0
19
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, merr family;PDBTitle: crystal structure of putative merr family transcriptional regulator2 from listeria monocytogenes
16 c2kfsA_
41.0
13
PDB header: dna-binding proteinChain: A: PDB Molecule: conserved hypothetical regulatory protein;PDBTitle: nmr structure of rv2175c
17 d1j9ia_
35.1
15
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Terminase gpNU1 subunit domain18 d2ezha_
26.0
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain19 c1b4aA_
25.3
22
PDB header: repressorChain: A: PDB Molecule: arginine repressor;PDBTitle: structure of the arginine repressor from bacillus stearothermophilus
20 d2ezia_
24.1
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain21 d1aoya_
not modelled
23.5
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain22 c3hosA_
not modelled
11.6
25
PDB header: transferase, dna binding protein/dnaChain: A: PDB Molecule: transposable element mariner, complete cds;PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
23 c3v4gA_
not modelled
10.2
9
PDB header: dna binding proteinChain: A: PDB Molecule: arginine repressor;PDBTitle: 1.60 angstrom resolution crystal structure of an arginine repressor2 from vibrio vulnificus cmcp6
24 d1pqwa_
not modelled
9.9
3
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain25 d1tvxb_
not modelled
9.9
38
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines26 c3l4hA_
not modelled
9.8
16
PDB header: protein bindingChain: A: PDB Molecule: e3 ubiquitin-protein ligase hecw1;PDBTitle: helical box domain and second ww domain of the human e3 ubiquitin-2 protein ligase hecw1
27 c1t6zB_
not modelled
9.7
18
PDB header: transferaseChain: B: PDB Molecule: riboflavin kinase/fmn adenylyltransferase;PDBTitle: crystal structure of riboflavin bound tm379
28 d1juqa_
not modelled
8.8
10
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain29 d1r6ra_
not modelled
8.5
22
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C30 c1r6rA_
not modelled
8.5
22
PDB header: viral proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: solution structure of dengue virus capsid protein reveals a2 new fold
31 c2dn4A_
not modelled
7.9
19
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i;PDBTitle: solution structure of rsgi ruh-060, a gtf2i domain in human2 cdna
32 c3hefB_
not modelled
7.8
31
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
33 c3eusB_
not modelled
7.8
9
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
34 d1sfka_
not modelled
7.7
22
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C35 c2d99A_
not modelled
7.7
19
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-048, a gtf2i domain in human2 cdna
36 d1b4aa1
not modelled
7.4
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain37 d1q60a_
not modelled
7.4
22
Fold: GTF2I-like repeatSuperfamily: GTF2I-like repeatFamily: GTF2I-like repeat38 c2ed2A_
not modelled
7.1
16
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i;PDBTitle: solution structure of rsgi ruh-069, a gtf2i domain in human2 cdna
39 d1tvxa_
not modelled
7.0
38
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines40 c2dn5A_
not modelled
7.0
25
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-057, a gtf2i domain in human2 cdna
41 c2dg6A_
not modelled
7.0
22
PDB header: gene regulationChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of the putative transcriptional regulator sco55502 from streptomyces coelicolor a3(2)
42 d1te7a_
not modelled
6.9
4
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: yqfB-like43 c2dzqA_
not modelled
6.8
16
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-066, a gtf2i domain in human2 cdna
44 c2e3lA_
not modelled
6.7
31
PDB header: transcriptionChain: A: PDB Molecule: transcription factor gtf2ird2 beta;PDBTitle: solution structure of rsgi ruh-068, a gtf2i domain in human2 cdna
45 c1iufA_
not modelled
6.6
25
PDB header: dna binding proteinChain: A: PDB Molecule: centromere abp1 protein;PDBTitle: low resolution solution structure of the two dna-binding2 domains in schizosaccharomyces pombe abp1 protein
46 d1f9na1
not modelled
6.4
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain47 d1f9pa_
not modelled
6.3
38
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines48 c2x0kB_
not modelled
6.0
21
PDB header: transferaseChain: B: PDB Molecule: riboflavin biosynthesis protein ribf;PDBTitle: crystal structure of modular fad synthetase from2 corynebacterium ammoniagenes
49 c2ejeA_
not modelled
5.4
16
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i;PDBTitle: solution structure of rsgi ruh-071, a gtf2i domain in human2 cdna
50 c2djyA_
not modelled
5.4
22
PDB header: ligase/signaling proteinChain: A: PDB Molecule: smad ubiquitination regulatory factor 2;PDBTitle: solution structure of smurf2 ww3 domain-smad7 py peptide2 complex
51 c2rn7A_
not modelled
5.1
36
PDB header: unknown functionChain: A: PDB Molecule: is629 orfa;PDBTitle: nmr solution structure of tnpe protein from shigella2 flexneri. northeast structural genomics target sfr125