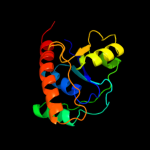
| 1 | c2fekA_ |
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: structure of a protein tyrosine phosphatase
|
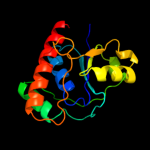
| 2 | c2wmyH_ |
|
 |
100.0 |
54 |
PDB header:hydrolase
Chain: H: PDB Molecule:putative acid phosphatase wzb;
PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
|
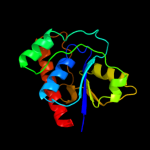
| 3 | d1dg9a_ |
|
 |
100.0 |
30 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
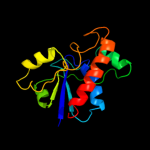
| 4 | c2gi4A_ |
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:possible phosphotyrosine protein phosphatase;
PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
|
| 5 | c3jviA_ |
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase;
PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
|
| 6 | c2cwdA_ |
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight phosphotyrosine protein phosphatase;
PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
|
| 7 | c3rofA_ |
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-phosphatase ptpa;
PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
|
| 8 | d5pnta_ |
|
 |
100.0 |
30 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 9 | d1p8aa_ |
|
 |
100.0 |
22 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 10 | c1zggA_ |
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative low molecular weight protein-tyrosine-
PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
|
| 11 | c1u2pA_ |
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
|
| 12 | d1d1qa_ |
|
 |
100.0 |
28 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 13 | c2l18A_ |
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:arsenate reductase;
PDBTitle: an arsenate reductase in the phosphate binding state
|
| 14 | d1jf8a_ |
|
 |
100.0 |
23 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 15 | d1jl3a_ |
|
 |
100.0 |
22 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 16 | d1y1la_ |
|
 |
100.0 |
27 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 17 | c3rh0A_ |
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:arsenate reductase;
PDBTitle: corynebacterium glutamicum mycothiol/mycoredoxin1-dependent arsenate2 reductase cg_arsc2
|
| 18 | c3t38B_ |
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:arsenate reductase;
PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
|
| 19 | c3fdfA_ |
|
 |
97.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:fr253;
PDBTitle: crystal structure of the serine phosphatase of rna2 polymerase ii ctd (ssu72 superfamily) from drosophila3 melanogaster. orthorhombic crystal form. northeast4 structural genomics consortium target fr253.
|
| 20 | c3o2qB_ |
|
 |
96.9 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
|
| 21 | c3o2sB_ |
|
not modelled |
96.6 |
32 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72 complex
|
| 22 | c2f00A_ |
|
not modelled |
93.3 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
| 23 | d1gpja2 |
|
not modelled |
91.8 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 24 | c1j6uA_ |
|
not modelled |
89.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase2 murc (tm0231) from thermotoga maritima at 2.3 a resolution
|
| 25 | d1p3da1 |
|
not modelled |
87.8 |
20 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 26 | c1gpjA_ |
|
not modelled |
86.7 |
16 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 27 | d1j6ua1 |
|
not modelled |
83.9 |
14 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 28 | c1gqqA_ |
|
not modelled |
82.6 |
20 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus2 influenzae
|
| 29 | c2bibA_ |
|
not modelled |
81.6 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:teichoic acid phosphorylcholine esterase/ choline binding
PDBTitle: crystal structure of the complete modular teichioic acid2 phosphorylcholine esterase pce (cbpe) from streptococcus3 pneumoniae
|
| 30 | c3eagA_ |
|
not modelled |
76.0 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-
PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
|
| 31 | c2eq8C_ |
|
not modelled |
72.3 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
| 32 | c3czcA_ |
|
not modelled |
71.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from2 streptococcus mutans
|
| 33 | d1vlva2 |
|
not modelled |
71.3 |
8 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 34 | d1u6ka1 |
|
not modelled |
71.3 |
25 |
Fold:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Superfamily:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Family:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) |
| 35 | d1vkra_ |
|
not modelled |
71.2 |
21 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 36 | c1vkrA_ |
|
not modelled |
71.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific pts system enzyme iiabc components;
PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
|
| 37 | c3ktdC_ |
|
not modelled |
71.0 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 38 | d1pjqa1 |
|
not modelled |
70.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 39 | c3kcqA_ |
|
not modelled |
69.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
|
| 40 | c3oj0A_ |
|
not modelled |
64.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: crystal structure of glutamyl-trna reductase from thermoplasma2 volcanium (nucleotide binding domain)
|
| 41 | c3hn7A_ |
|
not modelled |
63.8 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
|
| 42 | c2ph5A_ |
|
not modelled |
60.3 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
| 43 | c1e0cA_ |
|
not modelled |
57.3 |
16 |
PDB header:sulfurtransferase
Chain: A: PDB Molecule:sulfurtransferase;
PDBTitle: sulfurtransferase from azotobacter vinelandii
|
| 44 | c1wv9B_ |
|
not modelled |
56.3 |
28 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:rhodanese homolog tt1651;
PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
|
| 45 | d2czca2 |
|
not modelled |
56.2 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 46 | c3d1pA_ |
|
not modelled |
55.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative thiosulfate sulfurtransferase yor285w;
PDBTitle: atomic resolution structure of uncharacterized protein from2 saccharomyces cerevisiae
|
| 47 | c2k0zA_ |
|
not modelled |
55.7 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein hp1203;
PDBTitle: solution nmr structure of protein hp1203 from helicobacter pylori2 26695. northeast structural genomics consortium (nesg) target3 pt1/ontario center for structural proteomics target hp1203
|
| 48 | d1w4ha1 |
|
not modelled |
55.0 |
16 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
| 49 | d1yt8a4 |
|
not modelled |
54.8 |
12 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 50 | d1bala_ |
|
not modelled |
54.8 |
19 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
| 51 | d2z06a1 |
|
not modelled |
54.6 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
| 52 | d2fug21 |
|
not modelled |
54.5 |
29 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:NQO2-like |
| 53 | c3fojA_ |
|
not modelled |
53.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of ssp1007 from staphylococcus2 saprophyticus subsp. saprophyticus. northeast structural3 genomics target syr101a.
|
| 54 | d1w85i_ |
|
not modelled |
53.7 |
13 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
| 55 | c2i99A_ |
|
not modelled |
53.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mu-crystallin homolog;
PDBTitle: crystal structure of human mu_crystallin at 2.6 angstrom
|
| 56 | c1tvmA_ |
|
not modelled |
53.3 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
| 57 | c3c1oA_ |
|
not modelled |
52.4 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol synthase;
PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
|
| 58 | c2eq9C_ |
|
not modelled |
51.7 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdb
|
| 59 | d1wraa1 |
|
not modelled |
49.7 |
9 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Pce catalytic domain-like |
| 60 | c3dfzB_ |
|
not modelled |
49.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 61 | d1cmca_ |
|
not modelled |
49.5 |
15 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Met repressor, MetJ (MetR) |
| 62 | c2auvA_ |
|
not modelled |
49.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:potential nad-reducing hydrogenase subunit;
PDBTitle: solution structure of hndac : a thioredoxin-like [2fe-2s]2 ferredoxin involved in the nadp-reducing hydrogenase3 complex
|
| 63 | c2eq7C_ |
|
not modelled |
48.2 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:2-oxoglutarate dehydrogenase e2 component;
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdo
|
| 64 | d2cyua1 |
|
not modelled |
48.2 |
19 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
| 65 | c3tqrA_ |
|
not modelled |
48.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
| 66 | c3emeA_ |
|
not modelled |
46.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:rhodanese-like domain protein;
PDBTitle: crystal structure of rhodanese-like domain protein from2 staphylococcus aureus
|
| 67 | c1zwvA_ |
|
not modelled |
46.3 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:lipoamide acyltransferase component of branched-
PDBTitle: solution structure of the subunit binding domain (hbsbd) of2 the human mitochondrial branched-chain alpha-ketoacid3 dehydrogenase
|
| 68 | c1w3dA_ |
|
not modelled |
46.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dihydrolipoyllysine-residue acetyltransferase
PDBTitle: nmr structure of the peripheral-subunit binding domain of2 bacillus stearothermophilus e2p
|
| 69 | c2jrlA_ |
|
not modelled |
45.3 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
|
| 70 | c3i2vA_ |
|
not modelled |
44.9 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:adenylyltransferase and sulfurtransferase mocs3;
PDBTitle: crystal structure of human mocs3 rhodanese-like domain
|
| 71 | d1t71a_ |
|
not modelled |
43.6 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 72 | c1w4kA_ |
|
not modelled |
43.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate dehydrogenase e2;
PDBTitle: peripheral-subunit binding domains from mesophilic,2 thermophilic, and hyperthermophilic bacteria fold by3 ultrafast, apparently two-state transitions
|
| 73 | c3ilmD_ |
|
not modelled |
42.5 |
24 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:alr3790 protein;
PDBTitle: crystal structure of the alr3790 protein from anabaena sp. northeast2 structural genomics consortium target nsr437h
|
| 74 | c2e85B_ |
|
not modelled |
42.5 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase 3 maturation protease;
PDBTitle: crystal structure of the hydrogenase 3 maturation protease
|
| 75 | d1f0ya2 |
|
not modelled |
42.1 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 76 | c2cooA_ |
|
not modelled |
41.6 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:lipoamide acyltransferase component of branched-
PDBTitle: solution structure of the e3_binding domain of2 dihydrolipoamide branched chaintransacylase
|
| 77 | d1rrma_ |
|
not modelled |
41.0 |
7 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 78 | d1w85b2 |
|
not modelled |
41.0 |
11 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 79 | d1f37b_ |
|
not modelled |
40.8 |
22 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
| 80 | c2rgwD_ |
|
not modelled |
40.5 |
10 |
PDB header:transferase
Chain: D: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: catalytic subunit of m. jannaschii aspartate2 transcarbamoylase
|
| 81 | c3k96B_ |
|
not modelled |
38.7 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 82 | c3g5jA_ |
|
not modelled |
38.6 |
24 |
PDB header:nucleotide binding protein
Chain: A: PDB Molecule:putative atp/gtp binding protein;
PDBTitle: crystal structure of n-terminal domain of putative atp/gtp binding2 protein from clostridium difficile 630
|
| 83 | c2weuD_ |
|
not modelled |
36.9 |
20 |
PDB header:antifungal protein
Chain: D: PDB Molecule:tryptophan 5-halogenase;
PDBTitle: crystal structure of tryptophan 5-halogenase (pyrh) complex2 with substrate tryptophan
|
| 84 | c3dv0I_ |
|
not modelled |
36.6 |
18 |
PDB header:oxidoreductase/transferase
Chain: I: PDB Molecule:dihydrolipoyllysine-residue acetyltransferase
PDBTitle: snapshots of catalysis in the e1 subunit of the pyruvate2 dehydrogenase multi-enzyme complex
|
| 85 | d1jzta_ |
|
not modelled |
36.4 |
9 |
Fold:YjeF N-terminal domain-like
Superfamily:YjeF N-terminal domain-like
Family:YjeF N-terminal domain-like |
| 86 | d1okga2 |
|
not modelled |
35.4 |
9 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 87 | c3gt0A_ |
|
not modelled |
35.2 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: crystal structure of pyrroline 5-carboxylate reductase from bacillus2 cereus. northeast structural genomics consortium target bcr38b
|
| 88 | c3luyA_ |
|
not modelled |
34.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:probable chorismate mutase;
PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
|
| 89 | c3dcjA_ |
|
not modelled |
34.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable 5'-phosphoribosylglycinamide
PDBTitle: crystal structure of glycinamide formyltransferase (purn)2 from mycobacterium tuberculosis in complex with 5-methyl-5,3 6,7,8-tetrahydrofolic acid derivative
|
| 90 | d1cfza_ |
|
not modelled |
32.7 |
37 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Hydrogenase maturating endopeptidase HybD |
| 91 | d1urha1 |
|
not modelled |
32.6 |
21 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 92 | c3l6dB_ |
|
not modelled |
32.3 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 93 | d1e0ca1 |
|
not modelled |
32.2 |
16 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 94 | c2hi1A_ |
|
not modelled |
32.2 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
| 95 | c3bfjK_ |
|
not modelled |
31.5 |
12 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:1,3-propanediol oxidoreductase;
PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
|
| 96 | c3nbmA_ |
|
not modelled |
31.5 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
| 97 | d1iiba_ |
|
not modelled |
30.7 |
11 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 98 | c3ippA_ |
|
not modelled |
29.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:putative thiosulfate sulfurtransferase ynje;
PDBTitle: crystal structure of sulfur-free ynje
|
| 99 | d1e6ca_ |
|
not modelled |
29.0 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 100 | c2jtqA_ |
|
not modelled |
28.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phage shock protein e;
PDBTitle: rhodanese from e.coli
|
| 101 | d1umdb2 |
|
not modelled |
28.2 |
13 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 102 | d2jfga1 |
|
not modelled |
28.2 |
12 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 103 | d1ekxa2 |
|
not modelled |
28.0 |
13 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 104 | d2bw0a2 |
|
not modelled |
27.8 |
12 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 105 | c1s3iA_ |
|
not modelled |
26.5 |
12 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: crystal structure of the n terminal hydrolase domain of 10-2 formyltetrahydrofolate dehydrogenase
|
| 106 | d1meoa_ |
|
not modelled |
26.1 |
10 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 107 | c3gk5A_ |
|
not modelled |
26.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized rhodanese-related protein
PDBTitle: crystal structure of rhodanese-related protein (tvg0868615)2 from thermoplasma volcanium, northeast structural genomics3 consortium target tvr109a
|
| 108 | c1ulzA_ |
|
not modelled |
25.6 |
29 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase n-terminal domain;
PDBTitle: crystal structure of the biotin carboxylase subunit of pyruvate2 carboxylase
|
| 109 | d1dxha2 |
|
not modelled |
25.6 |
15 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 110 | d1t70a_ |
|
not modelled |
25.3 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 111 | d1duvg2 |
|
not modelled |
25.2 |
11 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 112 | c3nhzA_ |
|
not modelled |
24.9 |
27 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
| 113 | c3nhvE_ |
|
not modelled |
24.9 |
13 |
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:bh2092 protein;
PDBTitle: crystal structure of bh2092 protein from bacillus halodurans,2 northeast structural genomics consortium target bhr228f
|
| 114 | c3aaxB_ |
|
not modelled |
24.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:putative thiosulfate sulfurtransferase;
PDBTitle: crystal structure of probable thiosulfate sulfurtransferase2 cysa3 (rv3117) from mycobacterium tuberculosis: monoclinic3 form
|
| 115 | d1uara2 |
|
not modelled |
24.3 |
16 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 116 | c3cm0A_ |
|
not modelled |
24.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of adenylate kinase from thermus2 thermophilus hb8
|
| 117 | c3klbA_ |
|
not modelled |
23.7 |
30 |
PDB header:flavoprotein
Chain: A: PDB Molecule:putative flavoprotein;
PDBTitle: crystal structure of putative flavoprotein in complex with fmn2 (yp_213683.1) from bacteroides fragilis nctc 9343 at 1.75 a3 resolution
|
| 118 | d1rhsa1 |
|
not modelled |
23.6 |
13 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 119 | d2qmwa2 |
|
not modelled |
22.9 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 120 | c2dumD_ |
|
not modelled |
22.3 |
16 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ph0823;
PDBTitle: crystal structure of hypothetical protein, ph0823
|

