| 1 |
|
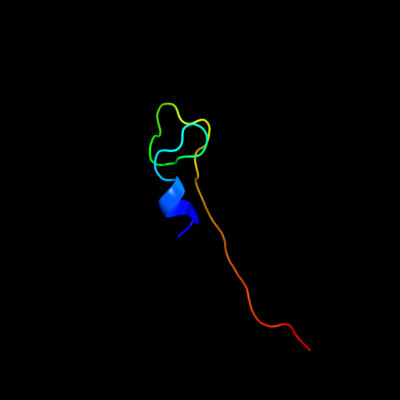
PDB 2wx6 chain B
Region: 8 - 43
Aligned: 29
Modelled: 36
Confidence: 40.1%
Identity: 41%
PDB header:oxidoreductase
Chain: B: PDB Molecule:peroxidase ycdb;
PDBTitle: x-ray crystallographic structure of e. coli apo-efeb
Phyre2
| 2 |
|
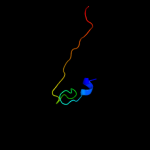
PDB 2iiz chain A domain 1
Region: 25 - 43
Aligned: 19
Modelled: 17
Confidence: 18.3%
Identity: 26%
Fold: Ferredoxin-like
Superfamily: Dimeric alpha+beta barrel
Family: Dyp-type peroxidase-like
Phyre2
| 3 |
|
PDB 1r2z chain A domain 3
Region: 33 - 40
Aligned: 8
Modelled: 8
Confidence: 9.2%
Identity: 50%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2
| 4 |
|
PDB 1k82 chain A domain 3
Region: 33 - 40
Aligned: 8
Modelled: 8
Confidence: 9.1%
Identity: 38%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2
| 5 |
|
PDB 1tdz chain A domain 3
Region: 33 - 41
Aligned: 9
Modelled: 9
Confidence: 8.7%
Identity: 56%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2
| 6 |
|
PDB 1k3x chain A domain 3
Region: 33 - 41
Aligned: 9
Modelled: 9
Confidence: 8.4%
Identity: 44%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2
| 7 |
|
PDB 1l1t chain A domain 3
Region: 33 - 40
Aligned: 8
Modelled: 8
Confidence: 8.3%
Identity: 50%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2
| 8 |
|
PDB 3qns chain A
Region: 25 - 43
Aligned: 19
Modelled: 19
Confidence: 7.6%
Identity: 21%
PDB header:oxidoreductase
Chain: A: PDB Molecule:dyp peroxidase;
PDBTitle: dypb from rhodococcus jostii rha1, crystal form 2
Phyre2
| 9 |
|
PDB 1ee8 chain A domain 3
Region: 33 - 41
Aligned: 9
Modelled: 9
Confidence: 7.5%
Identity: 56%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2