1 d2hjja1
100.0
100
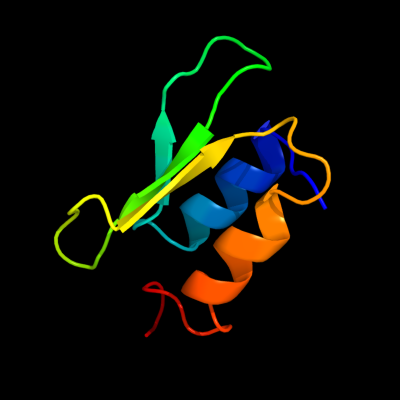
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: YkfF-like2 c2hjjA_
100.0
100
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ykff;PDBTitle: solution nmr structure of protein ykff from escherichia coli.2 northeast structural genomics target er397.
3 c3fcgB_
26.9
26
PDB header: membrane protein, protein transportChain: B: PDB Molecule: f1 capsule-anchoring protein;PDBTitle: crystal structure analysis of the middle domain of the2 caf1a usher
4 c3isyA_
22.9
36
PDB header: protein bindingChain: A: PDB Molecule: intracellular proteinase inhibitor;PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
5 c3hx8A_
19.2
29
PDB header: isomeraseChain: A: PDB Molecule: putative ketosteroid isomerase;PDBTitle: crystal structure of putative ketosteroid isomerase2 (np_103587.1) from mesorhizobium loti at 1.45 a resolution
6 c3ffeB_
17.2
31
PDB header: biosynthetic proteinChain: B: PDB Molecule: acsd;PDBTitle: structure of achromobactin synthetase protein d, (acsd)
7 c1xs3A_
16.3
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein xc975;PDBTitle: solution structure analysis of the xc975 protein
8 c2dhmA_
16.1
19
PDB header: protein bindingChain: A: PDB Molecule: protein bola;PDBTitle: solution structure of the bola protein from escherichia coli
9 d1xdna_
14.7
17
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: RNA ligase10 d1k1sa1
13.8
16
Fold: Lesion bypass DNA polymerase (Y-family), little finger domainSuperfamily: Lesion bypass DNA polymerase (Y-family), little finger domainFamily: Lesion bypass DNA polymerase (Y-family), little finger domain11 c2w02A_
13.0
31
PDB header: metal transportChain: A: PDB Molecule: acsd;PDBTitle: co-complex structure of achromobactin synthetase protein d (2 acsd) with atp from pectobacterium chrysanthemi
12 c3tr3A_
11.7
33
PDB header: unknown functionChain: A: PDB Molecule: bola;PDBTitle: structure of a bola protein homologue from coxiella burnetii
13 c1yewF_
11.1
50
PDB header: oxidoreductase, membrane proteinChain: F: PDB Molecule: particulate methane monooxygenase, a subunit;PDBTitle: crystal structure of particulate methane monooxygenase
14 c2flzC_
10.6
13
PDB header: hydrolaseChain: C: PDB Molecule: cis-3-chloroacrylic acid dehalogenase;PDBTitle: the x-ray structure of cis-3-chloroacrylic acid dehalogenase (cis-2 caad) with a sulfate ion bound in the active site
15 c3o2eA_
10.3
20
PDB header: unknown functionChain: A: PDB Molecule: bola-like protein;PDBTitle: crystal structure of a bol-like protein from babesia bovis
16 c2zt9F_
9.5
36
PDB header: photosynthesisChain: F: PDB Molecule: cytochrome b6-f complex subunit 7;PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
17 c2kdnA_
9.4
33
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein pfe0790c;PDBTitle: solution structure of pfe0790c, a putative bola-like2 protein from the protozoan parasite plasmodium falciparum.
18 c3lrmB_
9.2
23
PDB header: hydrolaseChain: B: PDB Molecule: alpha-galactosidase 1;PDBTitle: structure of alfa-galactosidase from saccharomyces cerevisiae with2 raffinose
19 c3chxF_
8.5
50
PDB header: membrane proteinChain: F: PDB Molecule: pmoa;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
20 d1f1sa4
8.0
21
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain21 c2f4qA_
not modelled
7.9
26
PDB header: isomeraseChain: A: PDB Molecule: type i topoisomerase, putative;PDBTitle: crystal structure of deinococcus radiodurans topoisomerase ib
22 d1ny8a_
not modelled
7.8
36
Fold: Alpha-lytic protease prodomain-likeSuperfamily: BolA-likeFamily: BolA-like23 c2h7fX_
not modelled
6.5
14
PDB header: isomerase/dnaChain: X: PDB Molecule: dna topoisomerase 1;PDBTitle: structure of variola topoisomerase covalently bound to dna
24 c2knrA_
not modelled
6.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atc0905;PDBTitle: solution structure of protein atu0922 from a. tumefaciens. northeast2 structural genomics consortium target att13. ontario center for3 structural proteomics target atc0905
25 d2oa4a1
not modelled
6.3
58
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: SPO1678-like26 d1v9ja_
not modelled
6.2
33
Fold: Alpha-lytic protease prodomain-likeSuperfamily: BolA-likeFamily: BolA-like27 c3iydB_
not modelled
6.2
19
PDB header: transcription/dnaChain: B: PDB Molecule: dna-directed rna polymerase subunit alpha;PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
28 d1x7fa2
not modelled
5.8
28
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Outer surface protein, N-terminal domain29 d1szwa_
not modelled
5.6
30
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: tRNA pseudouridine synthase TruD30 c1v60A_
not modelled
5.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: riken cdna 1810037g04;PDBTitle: solution structure of bola1 protein from mus musculus
31 d1ik0a_
not modelled
5.3
57
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Short-chain cytokines32 c2kz3A_
not modelled
5.3
28
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein rad51l3;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for human rad51d2 from 1 to 83
33 c2k17A_
not modelled
5.3
26
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation factor tfiid subunit 3;PDBTitle: solution structure of the taf3 phd domain in complex with a2 h3k4me3 peptide