| 1 |
|
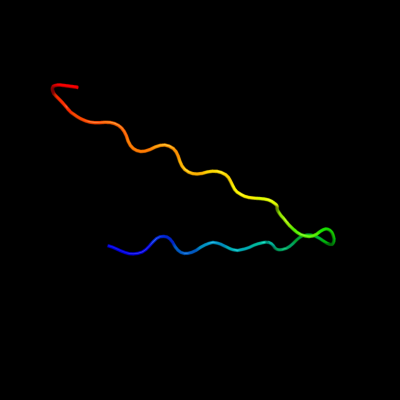
PDB 2ii8 chain F
Region: 44 - 67
Aligned: 24
Modelled: 24
Confidence: 56.5%
Identity: 29%
PDB header:signaling protein
Chain: F: PDB Molecule:anabaena sensory rhodopsin transducer protein;
PDBTitle: anabaena sensory rhodopsin transducer
Phyre2
| 2 |
|
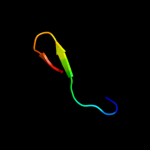
PDB 1nc7 chain A
Region: 43 - 68
Aligned: 26
Modelled: 26
Confidence: 55.5%
Identity: 27%
Fold: Hypothetical protein TM1070
Superfamily: Hypothetical protein TM1070
Family: Hypothetical protein TM1070
Phyre2
| 3 |
|
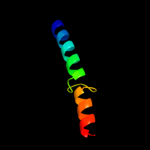
PDB 2krx chain A
Region: 43 - 57
Aligned: 15
Modelled: 15
Confidence: 11.1%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:asl3597 protein;
PDBTitle: solution nmr structure of asl3597 from nostoc sp. pcc7120. northeast2 structural genomics consortium target id nsr244.
Phyre2
| 4 |
|
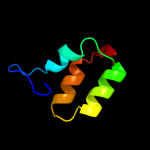
PDB 3arc chain L
Region: 27 - 42
Aligned: 16
Modelled: 16
Confidence: 8.7%
Identity: 44%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 5 |
|
PDB 3jrt chain A
Region: 32 - 42
Aligned: 11
Modelled: 11
Confidence: 6.3%
Identity: 36%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:integron cassette protein vpc_cass2;
PDBTitle: structure from the mobile metagenome of v. paracholerae:2 integron cassette protein vpc_cass2
Phyre2
| 6 |
|
PDB 1q90 chain M
Region: 12 - 48
Aligned: 31
Modelled: 37
Confidence: 5.6%
Identity: 39%
Fold: Single transmembrane helix
Superfamily: PetM subunit of the cytochrome b6f complex
Family: PetM subunit of the cytochrome b6f complex
Phyre2
| 7 |
|
PDB 2vvy chain C
Region: 1 - 51
Aligned: 47
Modelled: 50
Confidence: 5.4%
Identity: 30%
PDB header:viral protein
Chain: C: PDB Molecule:protein b15;
PDBTitle: structure of vaccinia virus protein b14
Phyre2