| 1 |
|
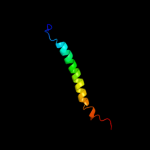
PDB 3cx5 chain D domain 2
Region: 338 - 378
Aligned: 39
Modelled: 41
Confidence: 14.9%
Identity: 18%
Fold: Single transmembrane helix
Superfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Family: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Phyre2
| 2 |
|
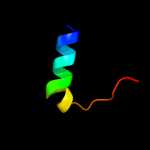
PDB 3ady chain A
Region: 361 - 373
Aligned: 13
Modelled: 13
Confidence: 8.9%
Identity: 31%
PDB header:proton transport
Chain: A: PDB Molecule:dotd;
PDBTitle: crystal structure of dotd from legionella
Phyre2
| 3 |
|
PDB 1ei5 chain A domain 2
Region: 366 - 378
Aligned: 13
Modelled: 13
Confidence: 6.6%
Identity: 38%
Fold: Streptavidin-like
Superfamily: D-aminopeptidase, middle and C-terminal domains
Family: D-aminopeptidase, middle and C-terminal domains
Phyre2
| 4 |
|
PDB 3geb chain C
Region: 362 - 374
Aligned: 12
Modelled: 13
Confidence: 6.5%
Identity: 50%
PDB header:hydrolase
Chain: C: PDB Molecule:eyes absent homolog 2;
PDBTitle: crystal structure of edeya2
Phyre2
| 5 |
|
PDB 1pzq chain A
Region: 364 - 385
Aligned: 22
Modelled: 22
Confidence: 5.9%
Identity: 36%
Fold: Dimerisation interlock
Superfamily: Docking domain A of the erythromycin polyketide synthase (DEBS)
Family: Docking domain A of the erythromycin polyketide synthase (DEBS)
Phyre2
| 6 |
|
PDB 1ppj chain D domain 2
Region: 338 - 376
Aligned: 37
Modelled: 37
Confidence: 5.8%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Family: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Phyre2
| 7 |
|
PDB 2knc chain A
Region: 329 - 385
Aligned: 43
Modelled: 43
Confidence: 5.6%
Identity: 9%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 8 |
|
PDB 2wwb chain B
Region: 301 - 318
Aligned: 18
Modelled: 18
Confidence: 5.5%
Identity: 22%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sec61 subunit gamma;
PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
Phyre2
| 9 |
|
PDB 3i71 chain B
Region: 363 - 385
Aligned: 23
Modelled: 23
Confidence: 5.4%
Identity: 30%
PDB header:unknown function
Chain: B: PDB Molecule:ethanolamine utilization protein eutk;
PDBTitle: ethanolamine utilization microcompartment shell subunit, eutk c-2 terminal domain
Phyre2