| 1 |
|
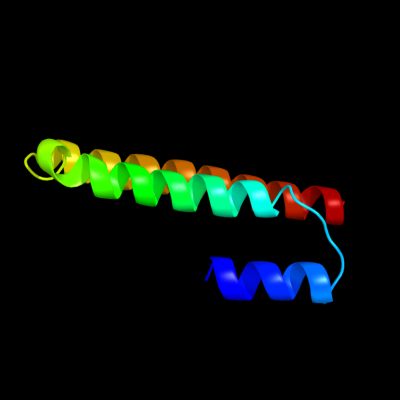
PDB 1oed chain E
Region: 7 - 79
Aligned: 73
Modelled: 73
Confidence: 21.8%
Identity: 23%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 2 |
|
PDB 2y69 chain Y
Region: 3 - 32
Aligned: 30
Modelled: 30
Confidence: 18.0%
Identity: 33%
PDB header:electron transport
Chain: Y: PDB Molecule:cytochrome c oxidase subunit 7c;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 3 |
|
PDB 1ehk chain B domain 2
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 17.4%
Identity: 39%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 4 |
|
PDB 1v54 chain L
Region: 3 - 32
Aligned: 30
Modelled: 30
Confidence: 15.7%
Identity: 33%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Family: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Phyre2
| 5 |
|
PDB 1ijd chain B
Region: 15 - 28
Aligned: 14
Modelled: 14
Confidence: 12.6%
Identity: 43%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 6 |
|
PDB 3l09 chain B
Region: 9 - 17
Aligned: 9
Modelled: 9
Confidence: 10.0%
Identity: 33%
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator2 (jann_22dec04_contig27_revised_gene3569) from jannaschia sp. ccs1 at3 2.81 a resolution
Phyre2
| 7 |
|
PDB 1nkz chain B
Region: 15 - 28
Aligned: 14
Modelled: 14
Confidence: 10.0%
Identity: 43%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 8 |
|
PDB 3koj chain A
Region: 69 - 81
Aligned: 13
Modelled: 13
Confidence: 9.0%
Identity: 23%
PDB header:dna binding protein
Chain: A: PDB Molecule:uncharacterized protein ycf41;
PDBTitle: crystal structure of the ssb domain of q5n255_synp6 protein2 from synechococcus sp. northeast structural genomics3 consortium target snr59a.
Phyre2
| 9 |
|
PDB 2wvq chain A
Region: 33 - 43
Aligned: 11
Modelled: 11
Confidence: 8.2%
Identity: 36%
PDB header:prion-binding protein
Chain: A: PDB Molecule:small s protein;
PDBTitle: structure of the het-s n-terminal domain. mutant d23a, p33h
Phyre2
| 10 |
|
PDB 3sqg chain E
Region: 20 - 44
Aligned: 25
Modelled: 25
Confidence: 7.6%
Identity: 36%
PDB header:transferase
Chain: E: PDB Molecule:methyl-coenzyme m reductase, beta subunit;
PDBTitle: crystal structure of a methyl-coenzyme m reductase purified from black2 sea mats
Phyre2
| 11 |
|
PDB 2pil chain A
Region: 25 - 39
Aligned: 14
Modelled: 15
Confidence: 5.8%
Identity: 50%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2