| 1 |
|
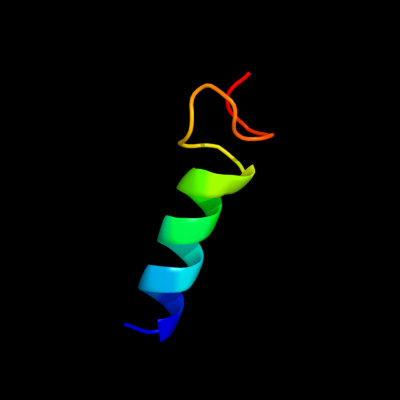
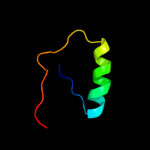
PDB 2b5d chain X domain 1
Region: 88 - 111
Aligned: 18
Modelled: 24
Confidence: 32.8%
Identity: 56%
Fold: immunoglobulin/albumin-binding domain-like
Superfamily: Families 57/38 glycoside transferase middle domain
Family: AmyC C-terminal domain-like
Phyre2
| 2 |
|
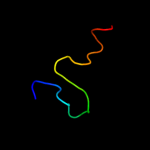
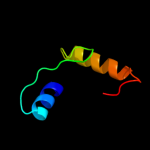
PDB 3bgh chain B
Region: 41 - 55
Aligned: 15
Modelled: 15
Confidence: 21.6%
Identity: 60%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative neuraminyllactose-binding hemagglutinin homolog;
PDBTitle: crystal structure of putative neuraminyllactose-binding hemagglutinin2 homolog from helicobacter pylori
Phyre2
| 3 |
|
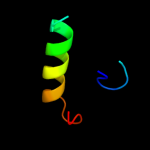
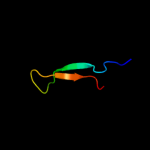
PDB 1ufa chain A domain 1
Region: 89 - 111
Aligned: 17
Modelled: 23
Confidence: 17.5%
Identity: 59%
Fold: immunoglobulin/albumin-binding domain-like
Superfamily: Families 57/38 glycoside transferase middle domain
Family: AmyC C-terminal domain-like
Phyre2
| 4 |
|
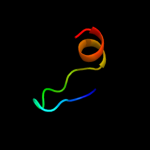
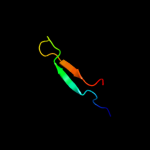
PDB 2i9i chain A
Region: 41 - 55
Aligned: 15
Modelled: 15
Confidence: 16.6%
Identity: 47%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of helicobacter pylori protein hp0492
Phyre2
| 5 |
|
PDB 2i9i chain A domain 1
Region: 41 - 55
Aligned: 15
Modelled: 15
Confidence: 16.6%
Identity: 47%
Fold: Anticodon-binding domain-like
Superfamily: XCC0632-like
Family: NLBH-like
Phyre2
| 6 |
|
PDB 2cu1 chain A domain 1
Region: 82 - 108
Aligned: 27
Modelled: 27
Confidence: 14.9%
Identity: 37%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 7 |
|
PDB 1jt6 chain A domain 2
Region: 41 - 67
Aligned: 27
Modelled: 27
Confidence: 12.8%
Identity: 30%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 8 |
|
PDB 2dlo chain A domain 2
Region: 73 - 90
Aligned: 18
Modelled: 18
Confidence: 12.2%
Identity: 22%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 9 |
|
PDB 2rbg chain B
Region: 52 - 86
Aligned: 33
Modelled: 35
Confidence: 12.0%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein st0493;
PDBTitle: crystal structure of hypothetical protein(st0493) from2 sulfolobus tokodaii
Phyre2
| 10 |
|
PDB 2npt chain B domain 1
Region: 64 - 108
Aligned: 45
Modelled: 45
Confidence: 11.1%
Identity: 27%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 11 |
|
PDB 1f2j chain A
Region: 11 - 35
Aligned: 25
Modelled: 25
Confidence: 6.6%
Identity: 36%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: Class I aldolase
Phyre2
| 12 |
|
PDB 3kx6 chain C
Region: 11 - 35
Aligned: 25
Modelled: 25
Confidence: 6.6%
Identity: 32%
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: crystal structure of fructose-1,6-bisphosphate aldolase from babesia2 bovis at 2.1a resolution
Phyre2
| 13 |
|
PDB 1xrx chain D
Region: 34 - 43
Aligned: 10
Modelled: 10
Confidence: 6.1%
Identity: 60%
PDB header:replication inhibitor
Chain: D: PDB Molecule:seqa protein;
PDBTitle: crystal structure of a dna-binding protein
Phyre2
| 14 |
|
PDB 1xrx chain A domain 1
Region: 34 - 43
Aligned: 10
Modelled: 10
Confidence: 6.1%
Identity: 60%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: SeqA N-terminal domain-like
Phyre2