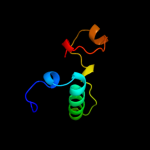
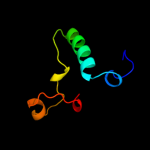
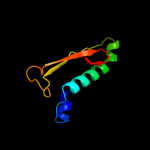
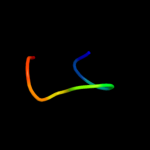1 d1gr0a1
24.0
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain2 c1fmeA_
22.3
53
PDB header: de novo proteinChain: A: PDB Molecule: fsd-ey peptide;PDBTitle: solution structure of fsd-ey, a novel peptide assuming a2 beta-beta-alpha fold
3 d2o62a1
19.9
11
Fold: LipocalinsSuperfamily: LipocalinsFamily: All1756-like4 c2ixsB_
16.4
19
PDB header: hydrolaseChain: B: PDB Molecule: sdai restriction endonuclease;PDBTitle: structure of sdai restriction endonuclease
5 c2e72A_
15.2
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pogo transposable element with znf domain;PDBTitle: solution structure of the zinc finger domain of human2 kiaa0461
6 d1tf3a1
14.2
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H27 d1tf6a1
10.7
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H28 d1uf2a_
10.1
27
Fold: Reovirus inner layer core protein p3Superfamily: Reovirus inner layer core protein p3Family: Phytoreovirus core9 c1uf2A_
10.1
27
PDB header: virusChain: A: PDB Molecule: core protein p3;PDBTitle: the atomic structure of rice dwarf virus (rdv)
10 c2k6rA_
10.1
56
PDB header: de novo proteinChain: A: PDB Molecule: full sequence design 1 synthetic superstable;PDBTitle: protein folding on a highly rugged landscape: experimental2 observation of glassy dynamics and structural frustration
11 d1q5qh_
10.0
10
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits12 c2h6jI_
9.3
10
PDB header: hydrolaseChain: I: PDB Molecule: proteasome beta-type subunit 1;PDBTitle: crystal structure of the beta f145a rhodococcus proteasome (casp2 target)
13 c1fsvA_
9.0
43
PDB header: beta beta alpha motifChain: A: PDB Molecule: full sequence design 1 of beta beta alpha motif;PDBTitle: full sequence design 1 (fsd-1) of beta beta alpha motif,2 nmr, minimized average structure
14 c1fsdA_
9.0
43
PDB header: novel sequenceChain: A: PDB Molecule: full sequence design 1 of beta beta alpha motif;PDBTitle: full sequence design 1 (fsd-1) of beta beta alpha motif,2 nmr, 41 structures
15 d1fnga2
8.5
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain16 d1jk8a2
8.1
21
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain17 c1mk2B_
8.0
46
PDB header: transcriptionChain: B: PDB Molecule: madh-interacting protein;PDBTitle: smad3 sbd complex
18 d1uvqa2
7.7
15
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain19 d1s9va2
7.6
21
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain20 c3gqiB_
7.6
22
PDB header: transferase/transferase inhibitorChain: B: PDB Molecule: phospholipase c-gamma-1;PDBTitle: crystal structure of activated receptor tyrosine kinase in2 complex with substrates
21 d1es0a2
not modelled
7.4
23
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain22 c1vd8A_
not modelled
7.3
55
PDB header: transcriptionChain: A: PDB Molecule: fibroin-modulator-binding-protein-1;PDBTitle: solution structure of fmbp-1 tandem repeat 2
23 c1wnmA_
not modelled
7.2
55
PDB header: transcriptionChain: A: PDB Molecule: fibroin-modulator binding-protein-1;PDBTitle: nmr structure of fmbp-1 tandem repeat 2 in 30%(v/v) tfe2 solution
24 d1xa6a2
not modelled
6.8
14
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain25 d2o0qa1
not modelled
6.6
73
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: CC0527-like26 c2pidB_
not modelled
6.3
30
PDB header: ligaseChain: B: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: crystal structure of human mitochondrial tyrosyl-trna synthetase in2 complex with an adenylate analog
27 d2gtlm1
not modelled
6.2
54
Fold: Streptavidin-likeSuperfamily: Extracellular hemoglobin linker subunit, receptor domainFamily: Extracellular hemoglobin linker subunit, receptor domain28 d1muja2
not modelled
6.1
15
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain29 d2p24a2
not modelled
5.5
15
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain30 d2gtlo1
not modelled
5.4
33
Fold: Streptavidin-likeSuperfamily: Extracellular hemoglobin linker subunit, receptor domainFamily: Extracellular hemoglobin linker subunit, receptor domain