| 1 |
|
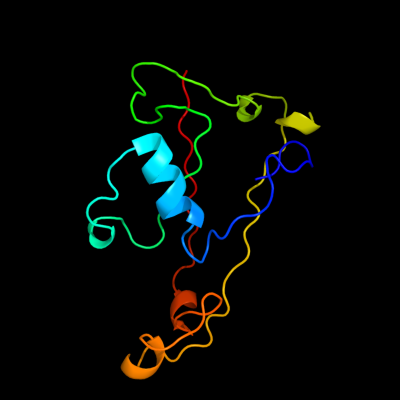
PDB 1j5q chain B
Region: 94 - 212
Aligned: 105
Modelled: 119
Confidence: 15.8%
Identity: 19%
PDB header:viral protein
Chain: B: PDB Molecule:major capsid protein;
PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
Phyre2
| 2 |
|
PDB 1f1s chain A domain 3
Region: 64 - 120
Aligned: 51
Modelled: 57
Confidence: 13.6%
Identity: 20%
Fold: Hyaluronate lyase-like, C-terminal domain
Superfamily: Hyaluronate lyase-like, C-terminal domain
Family: Hyaluronate lyase-like, C-terminal domain
Phyre2
| 3 |
|
PDB 1n7o chain A domain 2
Region: 58 - 124
Aligned: 61
Modelled: 67
Confidence: 11.9%
Identity: 20%
Fold: Hyaluronate lyase-like, C-terminal domain
Superfamily: Hyaluronate lyase-like, C-terminal domain
Family: Hyaluronate lyase-like, C-terminal domain
Phyre2
| 4 |
|
PDB 2x12 chain A
Region: 54 - 105
Aligned: 48
Modelled: 52
Confidence: 10.4%
Identity: 29%
PDB header:cell adhesion
Chain: A: PDB Molecule:fimbriae-associated protein fap1;
PDBTitle: ph-induced modulation of streptococcus parasanguinis2 adhesion by fap1 fimbriae
Phyre2
| 5 |
|
PDB 1bco chain A domain 1
Region: 106 - 123
Aligned: 18
Modelled: 18
Confidence: 8.2%
Identity: 22%
Fold: mu transposase, C-terminal domain
Superfamily: mu transposase, C-terminal domain
Family: mu transposase, C-terminal domain
Phyre2
| 6 |
|
PDB 2sh1 chain A
Region: 215 - 225
Aligned: 11
Modelled: 11
Confidence: 7.3%
Identity: 64%
Fold: Defensin-like
Superfamily: Defensin-like
Family: Defensin
Phyre2
| 7 |
|
PDB 1x3z chain A domain 1
Region: 114 - 147
Aligned: 34
Modelled: 34
Confidence: 7.1%
Identity: 15%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Transglutaminase core
Phyre2
| 8 |
|
PDB 1ulv chain A domain 3
Region: 125 - 158
Aligned: 31
Modelled: 34
Confidence: 6.7%
Identity: 35%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: CBD9-like
Family: Glucodextranase, domain C
Phyre2
| 9 |
|
PDB 2f4m chain A domain 1
Region: 114 - 156
Aligned: 43
Modelled: 43
Confidence: 6.0%
Identity: 16%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Transglutaminase core
Phyre2
| 10 |
|
PDB 2izp chain A domain 1
Region: 2 - 14
Aligned: 13
Modelled: 13
Confidence: 5.9%
Identity: 46%
Fold: IpaD-like
Superfamily: IpaD-like
Family: IpaD-like
Phyre2
| 11 |
|
PDB 2izp chain B
Region: 2 - 14
Aligned: 13
Modelled: 13
Confidence: 5.8%
Identity: 46%
PDB header:toxin
Chain: B: PDB Molecule:putative membrane antigen;
PDBTitle: bipd - an invasion prtein associated with the type-iii2 secretion system of burkholderia pseudomallei.
Phyre2
| 12 |
|
PDB 3m1h chain B
Region: 96 - 128
Aligned: 33
Modelled: 33
Confidence: 5.4%
Identity: 15%
PDB header:cell invasion
Chain: B: PDB Molecule:lysine specific cysteine protease;
PDBTitle: crystal structure analysis of the k3 cleaved adhesin domain of lys-2 gingipain (kgp) from porphyromonas gingivalis w83
Phyre2
| 13 |
|
PDB 1nvp chain D domain 2
Region: 128 - 147
Aligned: 20
Modelled: 20
Confidence: 5.2%
Identity: 25%
Fold: Transcription factor IIA (TFIIA), beta-barrel domain
Superfamily: Transcription factor IIA (TFIIA), beta-barrel domain
Family: Transcription factor IIA (TFIIA), beta-barrel domain
Phyre2