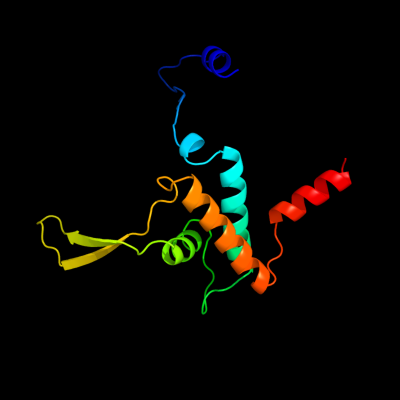
| 1 | d1pc6a_
|
|
 |
100.0 |
17 |
Fold:NinB
Superfamily:NinB
Family:NinB |
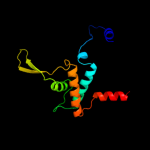
| 2 | c2kg4A_
|
|
 |
32.6 |
4 |
PDB header:cell cycle
Chain: A: PDB Molecule:growth arrest and dna-damage-inducible protein
PDBTitle: three-dimensional structure of human gadd45alpha in2 solution by nmr
|
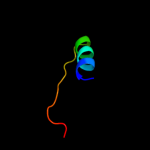
| 3 | c2jxjA_
|
|
 |
31.6 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:histone demethylase jarid1a;
PDBTitle: nmr structure of the arid domain from the histone h3k42 demethylase rbp2
|
| 4 | c2kngA_
|
|
 |
23.4 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein lsr2;
PDBTitle: solution structure of c-domain of lsr2
|
| 5 | c2lm1A_
|
|
 |
17.9 |
22 |
PDB header:dna binding protein
Chain: A: PDB Molecule:lysine-specific demethylase lid;
PDBTitle: solution nmr structure of lysine-specific demethylase lid from2 drosophila melanogaster, northeast structural genomics consortium3 target fr824d
|
| 6 | d2e1fa1
|
|
 |
16.7 |
10 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:HRDC domain from helicases |
| 7 | c3cg6A_
|
|
 |
14.8 |
6 |
PDB header:cell cycle
Chain: A: PDB Molecule:growth arrest and dna-damage-inducible 45 gamma;
PDBTitle: crystal structure of gadd45 gamma
|
| 8 | d1oi1a2
|
|
 |
13.8 |
27 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 9 | d1wh7a_
|
|
 |
12.0 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 10 | d1kkxa_
|
|
 |
11.5 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:ARID-like
Family:ARID domain |
| 11 | d1ax8a_
|
|
 |
11.4 |
15 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Long-chain cytokines |
| 12 | c1oz3C_
|
|
 |
11.2 |
43 |
PDB header:transcription
Chain: C: PDB Molecule:lethal(3)malignant brain tumor-like protein;
PDBTitle: crystal structure of 3-mbt repeats of lethal (3) malignant brain tumor2 (native-i) at 1.85 angstrom
|
| 13 | d1eysc_
|
|
 |
11.1 |
35 |
Fold:Multiheme cytochromes
Superfamily:Multiheme cytochromes
Family:Photosynthetic reaction centre (cytochrome subunit) |
| 14 | c1eysC_
|
|
 |
11.1 |
35 |
PDB header:electron transport
Chain: C: PDB Molecule:photosynthetic reaction center;
PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
|
| 15 | d1j20a1
|
|
 |
11.0 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 16 | d1bvp11
|
|
 |
11.0 |
19 |
Fold:A virus capsid protein alpha-helical domain
Superfamily:A virus capsid protein alpha-helical domain
Family:Orbivirus capsid |
| 17 | c3gwzB_
|
|
 |
10.9 |
7 |
PDB header:transferase
Chain: B: PDB Molecule:mmcr;
PDBTitle: structure of the mitomycin 7-o-methyltransferase mmcr
|
| 18 | d1oi1a1
|
|
 |
10.8 |
21 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 19 | d1jhga_
|
|
 |
10.6 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Trp repressor, TrpR |
| 20 | c2ztgA_
|
|
 |
9.8 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of archaeoglobus fulgidus alanyl-trna2 synthetase lacking the c-terminal dimerization domain in3 complex with ala-sa
|
| 21 | c2eqyA_ |
|
not modelled |
9.8 |
22 |
PDB header:dna binding protein
Chain: A: PDB Molecule:jumonji, at rich interactive domain 1b;
PDBTitle: solution structure of the arid domain of jarid1b protein
|
| 22 | d1wjra_ |
|
not modelled |
9.7 |
14 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 23 | d1wjsa_ |
|
not modelled |
9.3 |
23 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 24 | c3ut1A_ |
|
not modelled |
9.3 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:lethal(3)malignant brain tumor-like protein 3;
PDBTitle: crystal structure of the 3-mbt repeat domain of l3mbtl3
|
| 25 | c2yqeA_ |
|
not modelled |
9.2 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:jumonji/arid domain-containing protein 1d;
PDBTitle: solution structure of the arid domain of jarid1d protein
|
| 26 | d1oz2a2 |
|
not modelled |
9.0 |
43 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 27 | d2gtaa1 |
|
not modelled |
8.4 |
8 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 28 | c3frwF_ |
|
not modelled |
8.2 |
5 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative trp repressor protein;
PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
|
| 29 | d2cb2a1 |
|
not modelled |
7.8 |
21 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:SOR-like |
| 30 | d1miua4 |
|
not modelled |
7.7 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 31 | c2wvsD_ |
|
not modelled |
7.6 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 32 | c3lwzC_ |
|
not modelled |
7.5 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
|
| 33 | d2ahma1 |
|
not modelled |
7.3 |
0 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Coronavirus NSP7-like
Family:Coronavirus NSP7-like |
| 34 | d2gtad1 |
|
not modelled |
7.0 |
8 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 35 | d2aj6a1 |
|
not modelled |
6.9 |
18 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
| 36 | d1rfma_ |
|
not modelled |
6.8 |
13 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
| 37 | d1oz2a1 |
|
not modelled |
6.6 |
29 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 38 | d1trra_ |
|
not modelled |
6.4 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Trp repressor, TrpR |
| 39 | d1oz2a3 |
|
not modelled |
6.4 |
31 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 40 | c1yycA_ |
|
not modelled |
6.3 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative late embryogenesis abundant protein;
PDBTitle: solution structure of a putative late embryogenesis2 abundant (lea) protein at2g46140.1
|
| 41 | c3ebrA_ |
|
not modelled |
5.8 |
10 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized rmlc-like cupin;
PDBTitle: crystal structure of an rmlc-like cupin protein (reut_a0381) from2 ralstonia eutropha jmp134 at 2.60 a resolution
|
| 42 | d1hl9a2 |
|
not modelled |
5.8 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Putative alpha-L-fucosidase, catalytic domain |
| 43 | c1vl2C_ |
|
not modelled |
5.6 |
15 |
PDB header:ligase
Chain: C: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of argininosuccinate synthase (tm1780) from2 thermotoga maritima at 1.65 a resolution
|
| 44 | d1wjqa_ |
|
not modelled |
5.6 |
33 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 45 | d2ezha_ |
|
not modelled |
5.2 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |