| 1 |
|
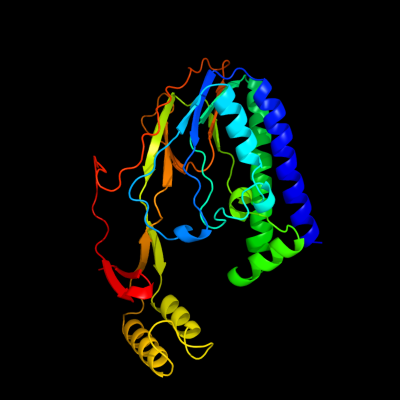
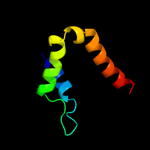
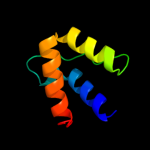
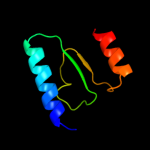
PDB 2i06 chain A domain 1
Region: 5 - 309
Aligned: 305
Modelled: 305
Confidence: 100.0%
Identity: 100%
Fold: Replication terminator protein (Tus)
Superfamily: Replication terminator protein (Tus)
Family: Replication terminator protein (Tus)
Phyre2
| 2 |
|
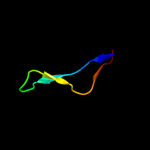
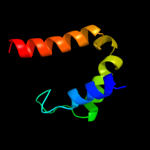
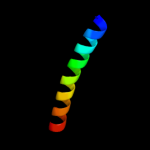
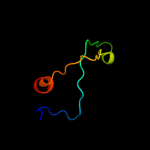
PDB 2a1j chain A domain 1
Region: 92 - 151
Aligned: 55
Modelled: 60
Confidence: 51.5%
Identity: 20%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Hef domain-like
Phyre2
| 3 |
|
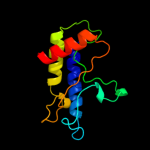
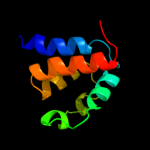
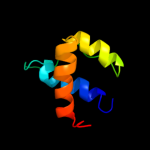
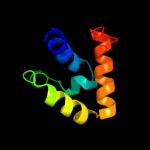
PDB 2gec chain A domain 1
Region: 232 - 306
Aligned: 71
Modelled: 75
Confidence: 33.7%
Identity: 18%
Fold: Coronavirus RNA-binding domain
Superfamily: Coronavirus RNA-binding domain
Family: Coronavirus RNA-binding domain
Phyre2
| 4 |
|
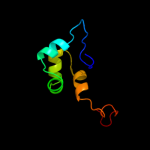
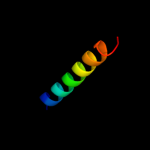
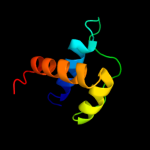
PDB 2bxx chain A domain 1
Region: 284 - 306
Aligned: 23
Modelled: 23
Confidence: 21.9%
Identity: 22%
Fold: Coronavirus RNA-binding domain
Superfamily: Coronavirus RNA-binding domain
Family: Coronavirus RNA-binding domain
Phyre2
| 5 |
|
PDB 2d3j chain A
Region: 154 - 176
Aligned: 23
Modelled: 23
Confidence: 18.7%
Identity: 13%
PDB header:signaling protein inhibitor
Chain: A: PDB Molecule:wnt inhibitory factor-1;
PDBTitle: nmr structure of the wif domain from human wif-1
Phyre2
| 6 |
|
PDB 3kx6 chain C
Region: 9 - 120
Aligned: 92
Modelled: 112
Confidence: 14.8%
Identity: 22%
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: crystal structure of fructose-1,6-bisphosphate aldolase from babesia2 bovis at 2.1a resolution
Phyre2
| 7 |
|
PDB 2aq0 chain A domain 1
Region: 92 - 158
Aligned: 62
Modelled: 67
Confidence: 14.8%
Identity: 18%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Hef domain-like
Phyre2
| 8 |
|
PDB 1vdd chain A
Region: 64 - 124
Aligned: 51
Modelled: 61
Confidence: 14.4%
Identity: 16%
Fold: Recombination protein RecR
Superfamily: Recombination protein RecR
Family: Recombination protein RecR
Phyre2
| 9 |
|
PDB 1vdd chain C
Region: 64 - 124
Aligned: 51
Modelled: 61
Confidence: 13.1%
Identity: 16%
PDB header:recombination
Chain: C: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recombinational repair protein recr
Phyre2
| 10 |
|
PDB 2km6 chain A
Region: 1 - 81
Aligned: 80
Modelled: 81
Confidence: 12.5%
Identity: 24%
PDB header:signaling protein, protein binding
Chain: A: PDB Molecule:nacht, lrr and pyd domains-containing protein 7;
PDBTitle: nmr structure of the nlrp7 pyrin domain
Phyre2
| 11 |
|
PDB 1ci6 chain B
Region: 8 - 31
Aligned: 24
Modelled: 24
Confidence: 11.4%
Identity: 25%
PDB header:transcription
Chain: B: PDB Molecule:transcription factor c/ebp beta;
PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
Phyre2
| 12 |
|
PDB 3qf2 chain B
Region: 18 - 81
Aligned: 63
Modelled: 64
Confidence: 11.3%
Identity: 19%
PDB header:apoptosis
Chain: B: PDB Molecule:nacht, lrr and pyd domains-containing protein 3;
PDBTitle: crystal structure of nalp3 pyd
Phyre2
| 13 |
|
PDB 2pnv chain A
Region: 4 - 31
Aligned: 28
Modelled: 28
Confidence: 10.5%
Identity: 25%
PDB header:membrane protein
Chain: A: PDB Molecule:small conductance calcium-activated potassium
PDBTitle: crystal structure of the leucine zipper domain of small-2 conductance ca2+-activated k+ (skca) channel from rattus3 norvegicus
Phyre2
| 14 |
|
PDB 1s1c chain Y
Region: 115 - 138
Aligned: 24
Modelled: 24
Confidence: 9.4%
Identity: 38%
PDB header:signaling protein
Chain: Y: PDB Molecule:rho-associated, coiled-coil containing protein
PDBTitle: crystal structure of the complex between the human rhoa and2 rho-binding domain of human rocki
Phyre2
| 15 |
|
PDB 1pn5 chain A domain 1
Region: 18 - 81
Aligned: 63
Modelled: 64
Confidence: 8.7%
Identity: 17%
Fold: DEATH domain
Superfamily: DEATH domain
Family: Pyrin domain, PYD
Phyre2
| 16 |
|
PDB 1pn5 chain A
Region: 18 - 81
Aligned: 63
Modelled: 64
Confidence: 8.7%
Identity: 17%
PDB header:apoptosis
Chain: A: PDB Molecule:nacht-, lrr- and pyd-containing protein 2;
PDBTitle: nmr structure of the nalp1 pyrin domain (pyd)
Phyre2
| 17 |
|
PDB 1a9x chain A domain 3
Region: 136 - 194
Aligned: 59
Modelled: 59
Confidence: 8.2%
Identity: 24%
Fold: PreATP-grasp domain
Superfamily: PreATP-grasp domain
Family: BC N-terminal domain-like
Phyre2
| 18 |
|
PDB 2gef chain A
Region: 225 - 267
Aligned: 42
Modelled: 43
Confidence: 7.3%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:protease vp4;
PDBTitle: crystal structure of a novel viral protease with a2 serine/lysine catalytic dyad mechanism
Phyre2
| 19 |
|
PDB 2do9 chain A
Region: 18 - 81
Aligned: 58
Modelled: 64
Confidence: 7.2%
Identity: 16%
PDB header:signaling protein
Chain: A: PDB Molecule:nacht-, lrr- and pyd-containing protein 10;
PDBTitle: solution structure of the pyrin/paad-dapin domain in mouse2 nalp10 (nacht, leucine rich repeat and pyd containing 10)
Phyre2