| 1 |
|
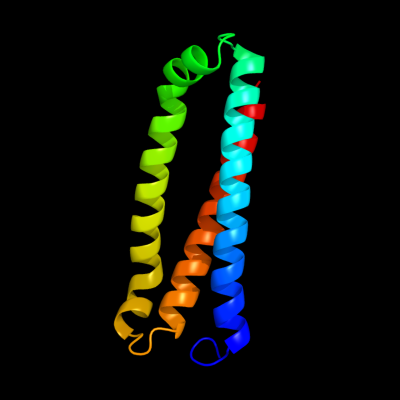
PDB 3gi7 chain A
Region: 42 - 152
Aligned: 103
Modelled: 111
Confidence: 99.9%
Identity: 15%
PDB header:unknown function
Chain: A: PDB Molecule:secreted protein of unknown function duf1311;
PDBTitle: crystal structure of a duf1311 family protein (pp0307) from2 pseudomonas putida kt2440 at 1.85 a resolution
Phyre2
| 2 |
|
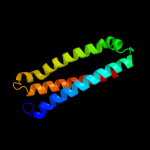
PDB 1n4c chain A
Region: 61 - 113
Aligned: 52
Modelled: 53
Confidence: 12.2%
Identity: 13%
Fold: Long alpha-hairpin
Superfamily: Chaperone J-domain
Family: Chaperone J-domain
Phyre2
| 3 |
|
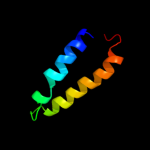
PDB 1zxa chain B
Region: 134 - 161
Aligned: 28
Modelled: 28
Confidence: 10.9%
Identity: 14%
PDB header:transferase
Chain: B: PDB Molecule:cgmp-dependent protein kinase 1, alpha isozyme;
PDBTitle: solution structure of the coiled-coil domain of cgmp-2 dependent protein kinase ia
Phyre2
| 4 |
|
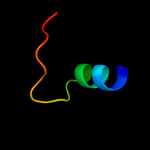
PDB 1pm6 chain A
Region: 142 - 162
Aligned: 21
Modelled: 21
Confidence: 9.2%
Identity: 14%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Excisionase-like
Phyre2
| 5 |
|
PDB 2rkh chain A
Region: 88 - 149
Aligned: 52
Modelled: 62
Confidence: 9.2%
Identity: 19%
PDB header:transcription
Chain: A: PDB Molecule:putative apha-like transcription factor;
PDBTitle: crystal structure of a putative apha-like transcription factor2 (zp_00208345.1) from magnetospirillum magnetotacticum ms-1 at 2.00 a3 resolution
Phyre2
| 6 |
|
PDB 2noc chain A domain 1
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 6.6%
Identity: 21%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 7 |
|
PDB 1rh6 chain A
Region: 142 - 162
Aligned: 21
Modelled: 21
Confidence: 6.5%
Identity: 14%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Excisionase-like
Phyre2
| 8 |
|
PDB 2jrm chain A
Region: 105 - 155
Aligned: 46
Modelled: 51
Confidence: 5.5%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribosome modulation factor;
PDBTitle: solution nmr structure of ribosome modulation factor vp1593 from2 vibrio parahaemolyticus. northeast structural genomics target vpr55
Phyre2
| 9 |
|
PDB 2dn9 chain A
Region: 61 - 123
Aligned: 59
Modelled: 63
Confidence: 5.2%
Identity: 14%
PDB header:apoptosis, chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily a member 3;
PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
Phyre2