| 1 |
|
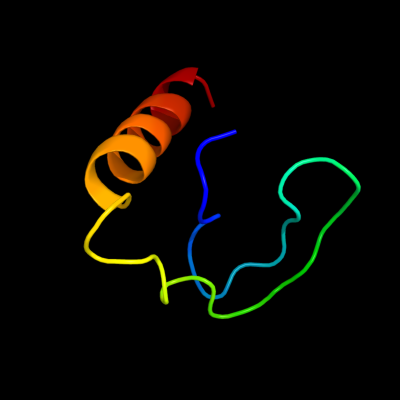
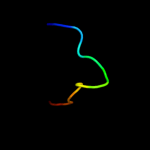
PDB 2d46 chain A
Region: 97 - 139
Aligned: 43
Modelled: 43
Confidence: 17.5%
Identity: 23%
PDB header:metal transport
Chain: A: PDB Molecule:calcium channel, voltage-dependent, beta 4
PDBTitle: solution structure of the human beta4a-a domain
Phyre2
| 2 |
|
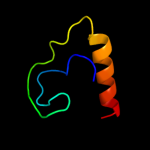
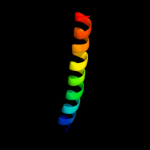
PDB 3i31 chain A
Region: 7 - 40
Aligned: 34
Modelled: 34
Confidence: 16.4%
Identity: 18%
PDB header:rna binding protein,hydrolase
Chain: A: PDB Molecule:heat resistant rna dependent atpase;
PDBTitle: hera helicase rna binding domain is an rrm fold
Phyre2
| 3 |
|
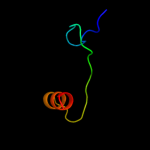
PDB 1x9b chain A
Region: 123 - 139
Aligned: 17
Modelled: 17
Confidence: 11.9%
Identity: 29%
Fold: Protozoan pheromone-like
Superfamily: Hypothetical membrane protein Ta0354, soluble domain
Family: Hypothetical membrane protein Ta0354, soluble domain
Phyre2
| 4 |
|
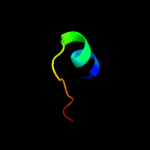
PDB 1ft8 chain E
Region: 72 - 83
Aligned: 12
Modelled: 12
Confidence: 10.5%
Identity: 33%
Fold: Ferredoxin-like
Superfamily: RNA-binding domain, RBD
Family: Non-canonical RBD domain
Phyre2
| 5 |
|
PDB 1zps chain A domain 1
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 8.6%
Identity: 38%
Fold: HisI-like
Superfamily: HisI-like
Family: HisI-like
Phyre2
| 6 |
|
PDB 2fef chain A domain 2
Region: 33 - 45
Aligned: 13
Modelled: 12
Confidence: 8.5%
Identity: 15%
Fold: PA2201 C-terminal domain-like
Superfamily: PA2201 C-terminal domain-like
Family: PA2201 C-terminal domain-like
Phyre2
| 7 |
|
PDB 1iml chain A domain 1
Region: 15 - 27
Aligned: 13
Modelled: 13
Confidence: 7.8%
Identity: 15%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 8 |
|
PDB 2cu8 chain A domain 1
Region: 15 - 27
Aligned: 13
Modelled: 13
Confidence: 7.3%
Identity: 15%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 9 |
|
PDB 2zjs chain E
Region: 28 - 57
Aligned: 30
Modelled: 30
Confidence: 7.1%
Identity: 20%
PDB header:protein transport/immune system
Chain: E: PDB Molecule:preprotein translocase sece subunit;
PDBTitle: crystal structure of secye translocon from thermus thermophilus with a2 fab fragment
Phyre2
| 10 |
|
PDB 2cql chain A domain 1
Region: 78 - 98
Aligned: 21
Modelled: 21
Confidence: 6.9%
Identity: 14%
Fold: Ribosomal protein L6
Superfamily: Ribosomal protein L6
Family: Ribosomal protein L6
Phyre2
| 11 |
|
PDB 3bm3 chain A
Region: 123 - 139
Aligned: 17
Modelled: 17
Confidence: 6.8%
Identity: 35%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:pspgi restriction endonuclease;
PDBTitle: restriction endonuclease pspgi-substrate dna complex
Phyre2