| 1 |
|
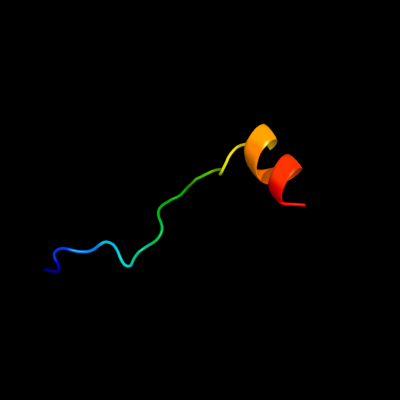
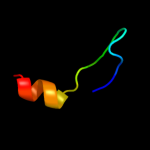
PDB 3bvh chain E
Region: 12 - 33
Aligned: 22
Modelled: 22
Confidence: 9.1%
Identity: 18%
PDB header:blood clotting
Chain: E: PDB Molecule:fibrinogen beta chain;
PDBTitle: crystal structure of recombinant gammad364a fibrinogen fragment d with2 the peptide ligand gly-pro-arg-pro-amide
Phyre2
| 2 |
|
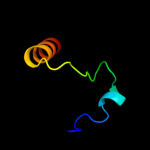
PDB 2rqx chain A
Region: 14 - 28
Aligned: 15
Modelled: 15
Confidence: 8.4%
Identity: 27%
PDB header:signaling protein
Chain: A: PDB Molecule:polymyxin b resistance protein;
PDBTitle: solution nmr structure of pmrd from klebsiella pneumoniae
Phyre2
| 3 |
|
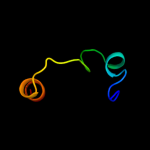
PDB 2ozl chain B domain 1
Region: 67 - 101
Aligned: 35
Modelled: 35
Confidence: 7.0%
Identity: 17%
Fold: Thiamin diphosphate-binding fold (THDP-binding)
Superfamily: Thiamin diphosphate-binding fold (THDP-binding)
Family: Branched-chain alpha-keto acid dehydrogenase Pyr module
Phyre2
| 4 |
|
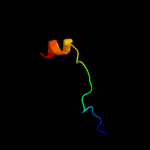
PDB 1qs0 chain B domain 1
Region: 67 - 101
Aligned: 35
Modelled: 32
Confidence: 6.5%
Identity: 9%
Fold: Thiamin diphosphate-binding fold (THDP-binding)
Superfamily: Thiamin diphosphate-binding fold (THDP-binding)
Family: Branched-chain alpha-keto acid dehydrogenase Pyr module
Phyre2
| 5 |
|
PDB 2bjo chain A
Region: 79 - 107
Aligned: 29
Modelled: 29
Confidence: 6.3%
Identity: 14%
PDB header:oxidoreductase
Chain: A: PDB Molecule:organic hydroperoxide resistance protein ohrb;
PDBTitle: crystal structure of the organic hydroperoxide resistance2 protein ohrb of bacillus subtilis
Phyre2
| 6 |
|
PDB 2hpc chain H
Region: 12 - 33
Aligned: 22
Modelled: 22
Confidence: 5.7%
Identity: 18%
PDB header:blood clotting
Chain: H: PDB Molecule:fibrinogen beta chain;
PDBTitle: crystal structure of fragment d from human fibrinogen complexed with2 gly-pro-arg-pro-amide.
Phyre2
| 7 |
|
PDB 2j61 chain B
Region: 14 - 33
Aligned: 20
Modelled: 20
Confidence: 5.4%
Identity: 20%
PDB header:lectin
Chain: B: PDB Molecule:ficolin-2;
PDBTitle: l-ficolin complexed to n-acetylglucosamine (forme c)
Phyre2