| 1 |
|
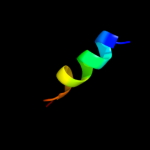
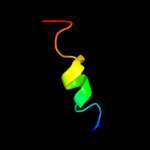
PDB 3lw5 chain K
Region: 63 - 78
Aligned: 16
Modelled: 16
Confidence: 17.1%
Identity: 44%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit x psak;
PDBTitle: improved model of plant photosystem i
Phyre2
| 2 |
|
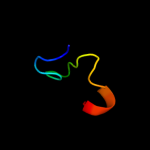
PDB 1e8g chain A domain 1
Region: 35 - 56
Aligned: 22
Modelled: 22
Confidence: 15.1%
Identity: 36%
Fold: Ferredoxin-like
Superfamily: FAD-linked oxidases, C-terminal domain
Family: Vanillyl-alcohol oxidase-like
Phyre2
| 3 |
|
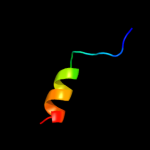
PDB 2hkv chain A domain 1
Region: 71 - 88
Aligned: 18
Modelled: 18
Confidence: 11.8%
Identity: 28%
Fold: DinB/YfiT-like putative metalloenzymes
Superfamily: DinB/YfiT-like putative metalloenzymes
Family: DinB-like
Phyre2
| 4 |
|
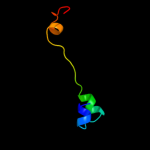
PDB 2q66 chain A domain 2
Region: 8 - 41
Aligned: 34
Modelled: 34
Confidence: 11.5%
Identity: 18%
Fold: Nucleotidyltransferase
Superfamily: Nucleotidyltransferase
Family: Poly(A) polymerase, PAP, N-terminal domain
Phyre2
| 5 |
|
PDB 3k13 chain A
Region: 28 - 59
Aligned: 30
Modelled: 31
Confidence: 10.9%
Identity: 20%
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
Phyre2
| 6 |
|
PDB 2cmp chain A
Region: 8 - 38
Aligned: 31
Modelled: 31
Confidence: 8.1%
Identity: 29%
PDB header:terminase
Chain: A: PDB Molecule:terminase small subunit;
PDBTitle: crystal structure of the dna binding domain of g1p small2 terminase subunit from bacteriophage sf6
Phyre2
| 7 |
|
PDB 1ahu chain B
Region: 35 - 56
Aligned: 22
Modelled: 22
Confidence: 6.9%
Identity: 36%
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
Phyre2
| 8 |
|
PDB 3c66 chain B
Region: 8 - 41
Aligned: 34
Modelled: 34
Confidence: 6.3%
Identity: 18%
PDB header:transferase
Chain: B: PDB Molecule:poly(a) polymerase;
PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
Phyre2
| 9 |
|
PDB 2g9l chain A
Region: 25 - 47
Aligned: 23
Modelled: 23
Confidence: 5.7%
Identity: 43%
PDB header:antibiotic
Chain: A: PDB Molecule:gaegurin-4;
PDBTitle: the high-resolution solution conformation of an2 antimicrobial peptide gaegurin 4 and its mode of membrane3 interaction
Phyre2
| 10 |
|
PDB 2wsf chain G
Region: 63 - 78
Aligned: 16
Modelled: 16
Confidence: 5.3%
Identity: 44%
PDB header:photosynthesis
Chain: G: PDB Molecule:photosystem i reaction center subunit v,
PDBTitle: improved model of plant photosystem i
Phyre2