Missense3D server is freely available to the scientific community.
1. Type the PDB coordinate on the file of interest e.g. 2fos or upload a 3D coordinate file, either by specifying a PDB code or by providing your own 3D coordinate set.
2. Specify the residue number. Warning: sequence numbering in PDB files may not correspond to that reported in UniProt (e.g. Residue Cys 56 in the Alpha-galactosidase protein (UniProt ID: P06280) corresponds to Cys 52 in PDB file 3hg3.pdb).
3. Specify the chain identifier in your 3D coordinate file. Warning: several 3D prediction servers like our in house Phyre2 do not add a chain identifier in the 3D coordinate file of the model. If your 3D coordinate file does not have a chain id, plese put a dash symbol "-" in this field.
4. Specify the wild type residue and its substitution.
5. Press button
6. If you have uploaded the wrong 3D coordinate file or simply want to reset the whole page, press the RESET button
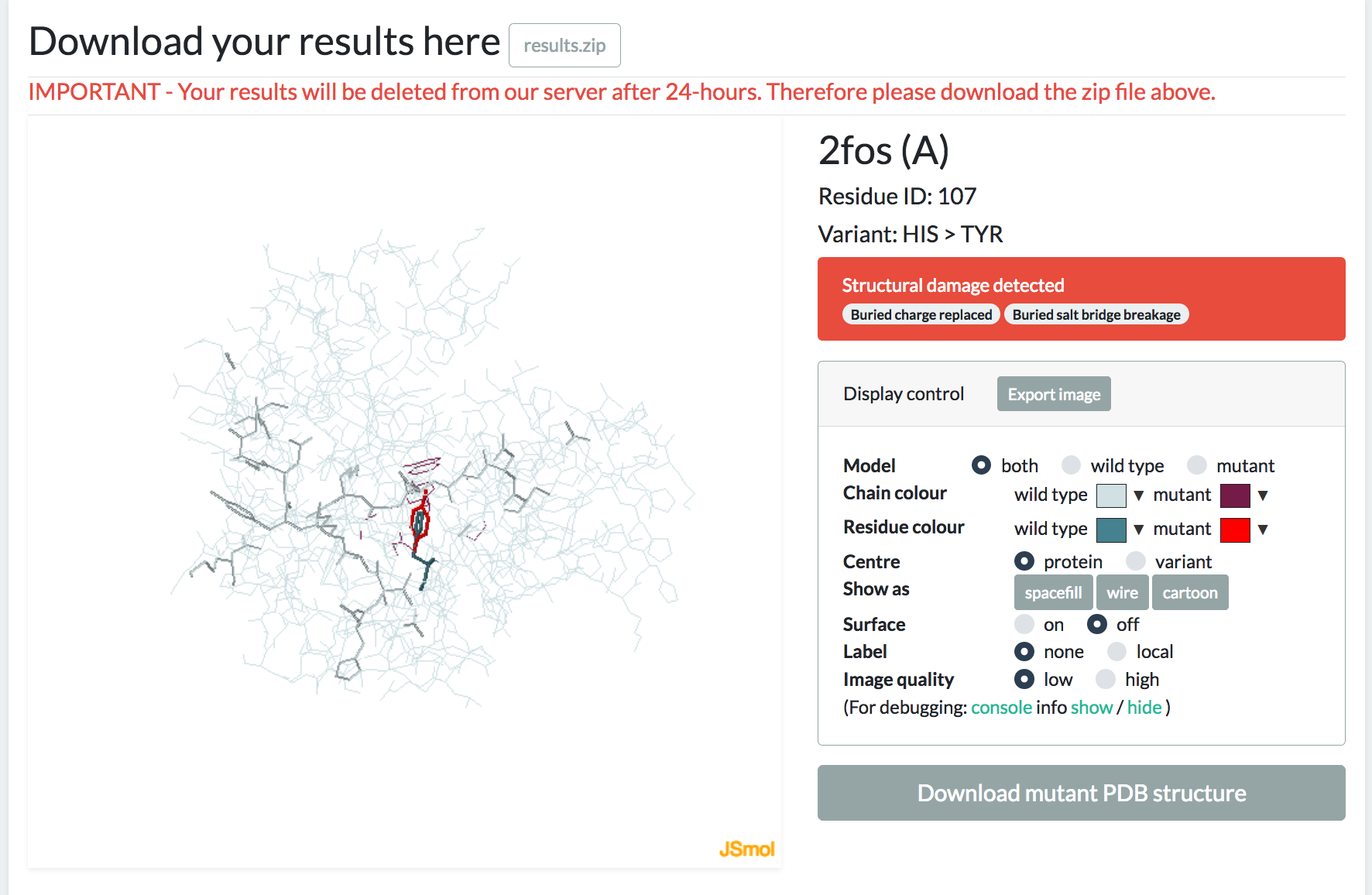
The output from Missense3D is generally produced within 3 minutes. A link to the result page will become available once the job is finished. The link will take you to a concise report of the structural changes identified by Missense3D. All results can be downloaded as a zip file from this page. Please note your results will be deleted from our server after 24-hours. Therefore please download the zip file if you want to save your results.
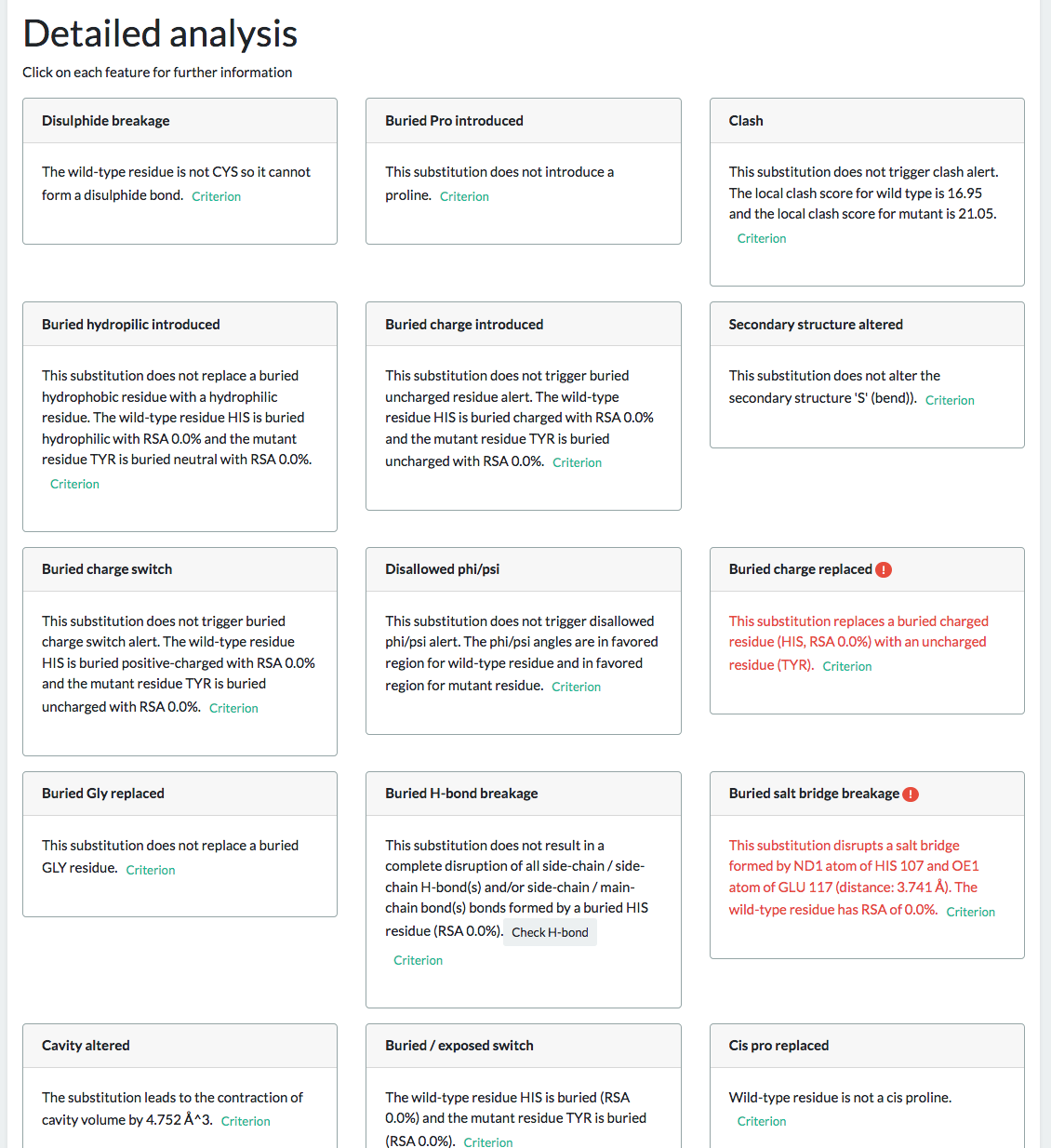
The “View Details” link will take you to a more detailed page (all results can also be download as a zip file from this page)