
 | Help |
|
| From protein 3D structure to Gene Ontology (GO) terms | ||
| 3d2GO Search - Help - Contact - Disclaimer - Example | ||
3d2GO uses several methods of function prediction, using sequence and structure, to predict Gene Ontology (GO) terms for your protein. The information from these various methods constitutes the input to a machine learning algorithm called a Support Vector Machine (SVM). Using a benchmark of diverse proteins with known GO annotations, the SVM has been trained to discriminate between true and false positive annotations based on the outputs of the set of algorithms used in 3D2GO.
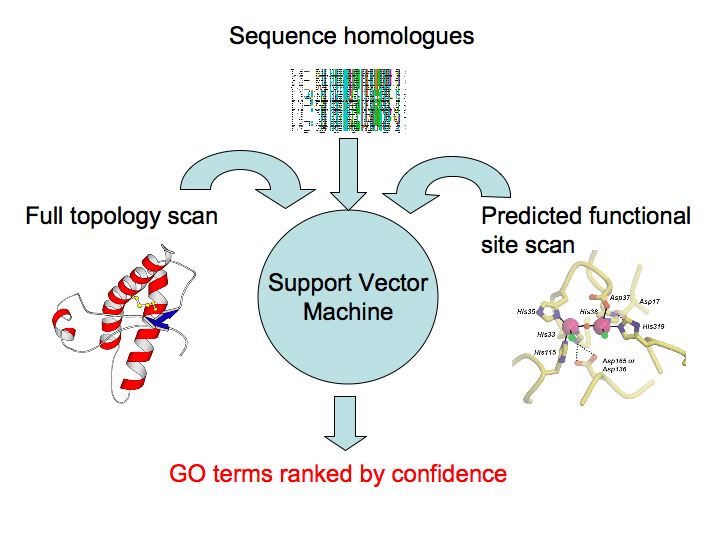
Full Topology SearchA full structural scan of your protein structure is made against the Structural Classification of Proteins (SCOP) database using a modified version of BLAST [1]. This converts your structure into a a string using a structural alphabet. This string is searched using the conventional BLAST algorithm against a database of pre-compiled structural sequences. In addition, the slower but more sensitive MAMMOTH [2]structural alignment program is used. Confident hits to known structures are stored, together with their associated GO terms. Functional Site PredictionPSI-Blast [3] is used to search for homologues of your protein sequence. These homologues are realigned using MUSCLE [4]. This alignment is used to perform a functional residue prediction using the Jenson-Shannon Divergence [5]. This is an information-theory approach to determine relative residue conservation. Such conservation is related to the functional importance of residues. Pocket/cleft DetectionDeep pockets or clefts in a protein are frequently associated with biological function. We detect such pockets by calculating the convex hull of a protein structure and its Delauney triangulation [6]. By calculating the conservation score of each pocket it is possible to rank the pockets for predicted functional relevance as well as the residues within each pocket. Functional Site SearchGiven candidate functional site residues, one may scan such sites against structures of known function using a fast geometric hashing technique [7]. This permits one to detect similar 3D patterns of residues, irrespective of connectivity, within other structures and rank potential matches by the quality of superposition and similarity of composition. Sequence HomologyUsing weak similarity to functionally annotated protein sequences in conjunction with the statistical relationships between the rate of co-occurrence of different biological functions has been shown to enhance functional annotation in the absence of clear homology [8]. Support Vector MachineSVMs are machine learning devices that allow one to quickly and accurately solve discrimination (and other) problems in the face of normally intractable high dimensional non-linear search spaces. We combine the confidence values, frequency of hits and background frequencies of GO terms from the above methods into a feature vector. Having trained an SVM on a benchmark of known structures, this vector can be classified as a true or false annotation. The output of the SVM can then be recast as a probability by fitting a probability distribution to training data. This final probability is that presented in the summary of results from a 3D2GO run. References1. C.-H. Tung, J.-W. Huang and J.-M. Yang "Kappa-alpha plot derived structural alphabet and BLOSUM-like substitution matrix for fast protein structure database search," Genome Biology, vol. 8, pp. R31.1~R31.16, 2007. 2. Ortiz, A. R., C. E. Strauss, and O. Olmea (2002, November). Mammoth (matching molecular models obtained from theory): An automated method for model comparison. Protein Sci 11 (11), 2606-2621. 3. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. SF Altschul, TL Madden, AA Schaffer, J Zhang, Z Zhang, W Miller and DJ Lipman. Nucleic Acids Research, Vol 25, Issue 17 3389-3402. 1997 4. Edgar, Robert C. (2004), MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Research 32(5), 1792-97. 5. Capra J, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics (2007) 23:1875 6. H. Edelsbrunner and M.Facello and Jie Liang. (1998). On the definition and the construction of pockets in macromolecules. [http://dx.doi.org/10.1016/S0166-218X(98)00067-5]. Discrete and Applied Mathematics, 88, 83-102. 7. M. Moll and L.E. Kavraki. Matching of Structural Motifs Using Hashing on Residue Labels and Geometric Filtering for Protein Function Prediction. The Seventh Annual International Conference on Computational Systems Bioinformatics (CSB2008), Stanford, CA, 2008. 8. Hawkins, T., Luban, S. and Kihara, D. 2006. Enhanced Automated Function Prediction Using Distantly Related Sequences and Contextual Association by PFP. Protein Science 15: 1550-6. A typical results screen is shown below. Explanations of the screenshot follow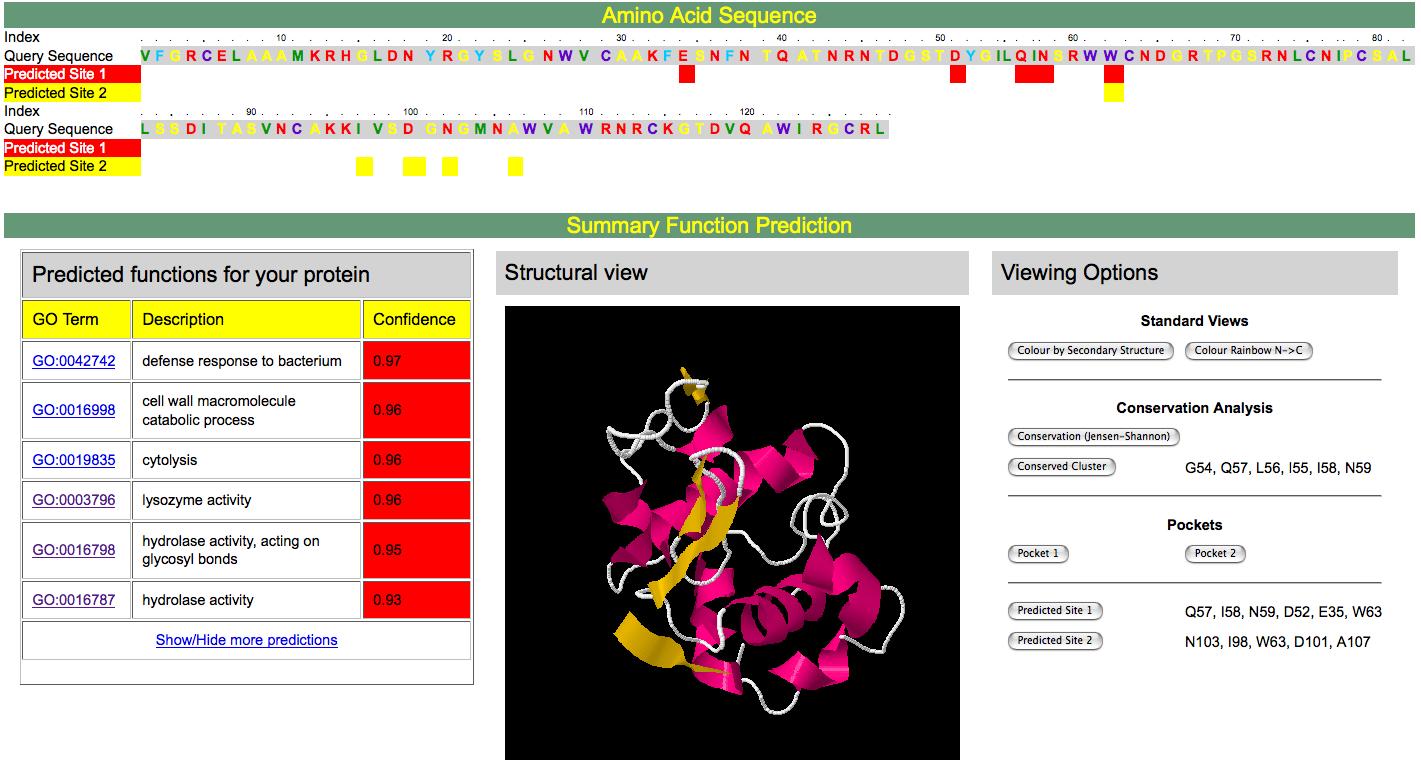
Amino Acid SequenceThis section shows the sequence of the user-submitted structure, and potentially functional important sites in yellow and red. These highlighted residues are used in the local structure matching part of the algorithm. Summary Function PredictionThis section is divided into three subsections. On the left, are the predicted GO terms for the structure, their descriptions from GO and a confidence score. Confidence ranges from 0 to 1, with 1 being the most confident prediction. High confidence predictions are shown towards the red end of the rainbow. Below this table is a link to display more predictions. By default only the top 6 predictions are shown. The middle window is a JMOL applet permitting the user to examine their structure - the JAVA runtime environment is required for this and is freely avalable from www.java.com if it is not already installed on your system. Documentation on using JMOL can be found at www.jmol.org The rightmost panel provides the user with a range of viewing options to highlight the results of the functional site predictions, pockets, Jensen-Shannon divergence, and the residue indices of the predicted active sites. Finally, not shown in this screen shot, there is a link below this information that permits a detailed look at the outputs of all the individual algorithms and the associated individual confidence scores before processing by the Support Vector Machine. Lawrence Kelley
| |||||||