|
ConFunc Example Submission
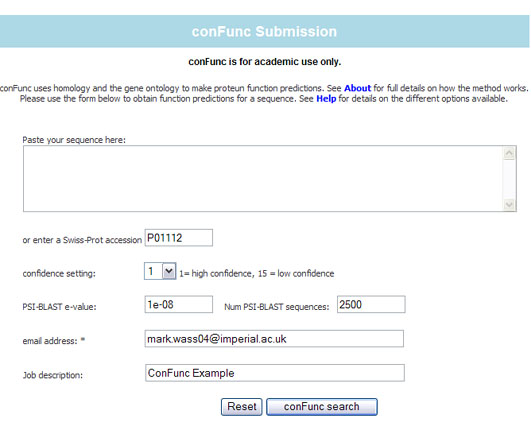
In this example:
- a Swiss-Prot accession has been entered rather than pasting a sequence.
- Confidence is set to 1 - so only GO terms with greatest confidence will be predicted
- PSI-BLAST e-value - default has been used. Enter a different value to modify this setting.
- increasing this value may result in identifying more annotated homologues for the query sequence.
- reducing the value will reduce the number of sequences identified
- PSI-BLAST number of homologues - set the maximum number of homologues that PSI-BLAST will return.
- email address -an email address is essential so that ConFunc can inform you of progress with your job
- each job is run on a computing farm system. If there are many jobs runnning then there may
be a delay in the job running.
- A description is optional - so you can identify the job.
Now view a breakdown of the results for this run
|

