|
ConFunc Example Results
view the full results page for this example: http://www.sbg.bio.ic.ac.uk/confunc/qconfunc_report.cgi?jobid=example
or work through the explanation of the results page below.
The Top of The Results Page
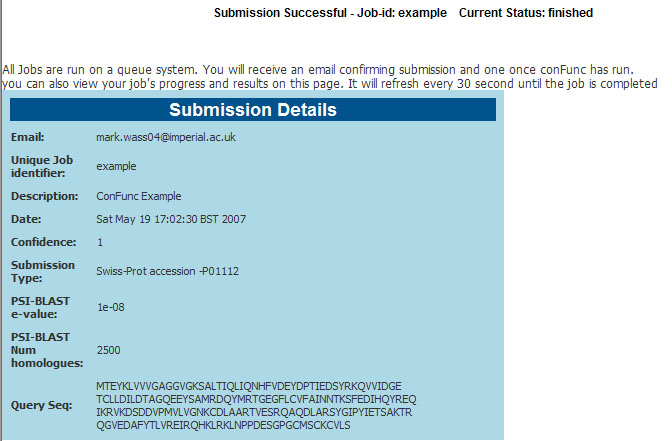
- As your job runs the results page will continue to be updated. At the top of the page the status is shown.
- The submission details are also printed for information as shown in the figure below.
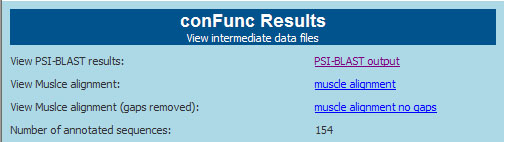
Results Data Files
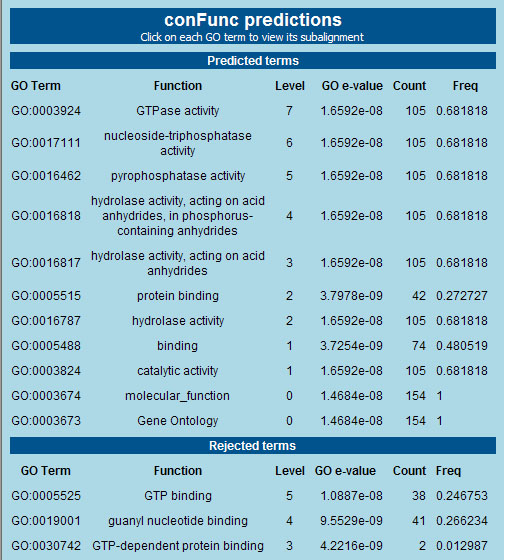
GO Predictions
- Finally on the results page the predicted and rejected GO terms are displayed. The predicted terms are shown first.
- For each GO term the following are displayed:
- a description of its function
- its level (with Molecular function considered to be level 0)
- a GO term c-value generated by ConFunc
- the count - absolute number of time GO term present in set of homologues
- frequency - percentage presence of GO term in set of annotated homologues
Some of the results for the example are shown below:
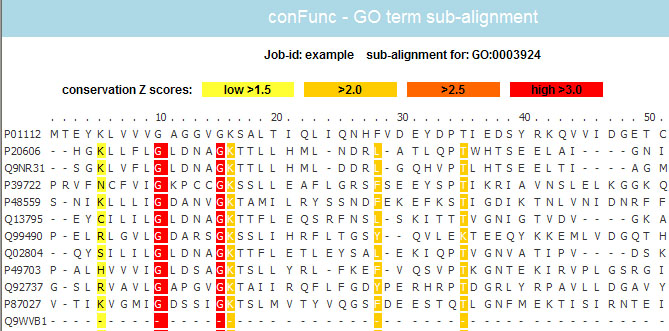
GO term sub-alignments
A sub-alignment is associated with each GO term. Simply click on a term is display its sub-alignment. So you would click on GO:0003924 in the above example to view its sub-alignment.
A portion of this sub-alignment is shown below.
- Functional residues identified by ConFunc are coloured.
- The key at the top describes the colouring scheme. The darker the colour the more conserved it is.
view the full results page for this example: http://www.sbg.bio.ic.ac.uk/confunc/qconfunc_report.cgi?jobid=example
|

