| 1 |
|
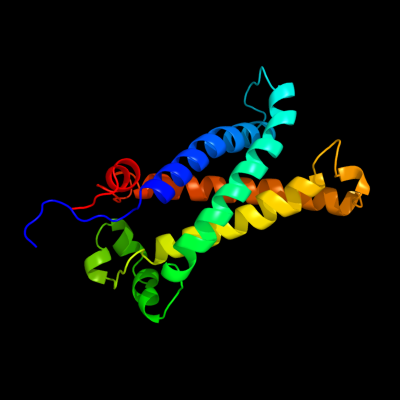
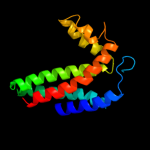
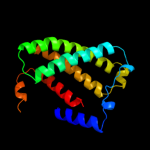
PDB 1kqf chain C
Region: 61 - 260
Aligned: 183
Modelled: 187
Confidence: 100.0%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Formate dehydrogenase N, cytochrome (gamma) subunit
Phyre2
| 2 |
|
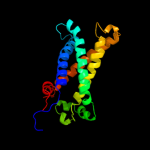
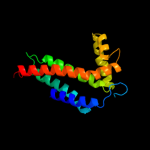
PDB 1bcc chain C domain 3
Region: 79 - 241
Aligned: 147
Modelled: 151
Confidence: 97.8%
Identity: 14%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 3 |
|
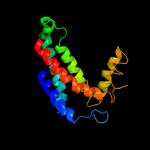
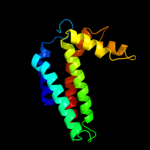
PDB 1ppj chain C domain 2
Region: 79 - 241
Aligned: 147
Modelled: 147
Confidence: 97.8%
Identity: 15%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 4 |
|
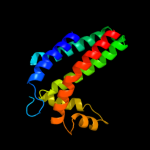
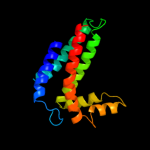
PDB 3cwb chain C
Region: 79 - 240
Aligned: 146
Modelled: 146
Confidence: 97.8%
Identity: 13%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
Phyre2
| 5 |
|
PDB 1q90 chain B
Region: 79 - 249
Aligned: 155
Modelled: 155
Confidence: 97.6%
Identity: 14%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 6 |
|
PDB 3cx5 chain C domain 2
Region: 79 - 241
Aligned: 147
Modelled: 147
Confidence: 97.6%
Identity: 12%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 7 |
|
PDB 3cx5 chain N
Region: 79 - 240
Aligned: 146
Modelled: 146
Confidence: 97.5%
Identity: 12%
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
Phyre2
| 8 |
|
PDB 2e74 chain A domain 1
Region: 79 - 241
Aligned: 149
Modelled: 151
Confidence: 97.5%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 9 |
|
PDB 2qjk chain M
Region: 79 - 241
Aligned: 147
Modelled: 147
Confidence: 97.5%
Identity: 13%
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
Phyre2
| 10 |
|
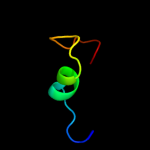
PDB 1kqf chain B domain 2
Region: 31 - 62
Aligned: 32
Modelled: 32
Confidence: 46.6%
Identity: 9%
Fold: Single transmembrane helix
Superfamily: Iron-sulfur subunit of formate dehydrogenase N, transmembrane anchor
Family: Iron-sulfur subunit of formate dehydrogenase N, transmembrane anchor
Phyre2
| 11 |
|
PDB 2knc chain A
Region: 22 - 65
Aligned: 44
Modelled: 44
Confidence: 20.3%
Identity: 11%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 12 |
|
PDB 3mk7 chain F
Region: 29 - 103
Aligned: 71
Modelled: 75
Confidence: 9.6%
Identity: 17%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 13 |
|
PDB 1y5i chain C domain 1
Region: 40 - 235
Aligned: 153
Modelled: 155
Confidence: 8.1%
Identity: 13%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 14 |
|
PDB 1ji8 chain A
Region: 2 - 26
Aligned: 25
Modelled: 25
Confidence: 7.6%
Identity: 32%
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily: DsrC, the gamma subunit of dissimilatory sulfite reductase
Family: DsrC, the gamma subunit of dissimilatory sulfite reductase
Phyre2
| 15 |
|
PDB 2kxa chain A
Region: 248 - 259
Aligned: 12
Modelled: 12
Confidence: 7.3%
Identity: 25%
PDB header:viral protein, immune system
Chain: A: PDB Molecule:haemagglutinin ha2 chain peptide;
PDBTitle: the hemagglutinin fusion peptide (h1 subtype) at ph 7.4
Phyre2
| 16 |
|
PDB 1fft chain G
Region: 29 - 89
Aligned: 59
Modelled: 61
Confidence: 7.1%
Identity: 17%
PDB header:oxidoreductase
Chain: G: PDB Molecule:ubiquinol oxidase;
PDBTitle: the structure of ubiquinol oxidase from escherichia coli
Phyre2