| 1 |
|
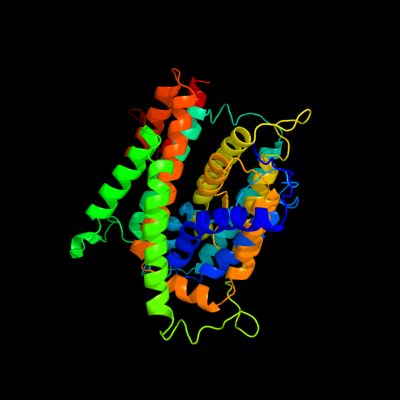
PDB 3qe7 chain A
Region: 22 - 459
Aligned: 399
Modelled: 424
Confidence: 100.0%
Identity: 25%
PDB header:transport protein
Chain: A: PDB Molecule:uracil permease;
PDBTitle: crystal structure of uracil transporter--uraa
Phyre2
| 2 |
|
PDB 1bzk chain A
Region: 29 - 62
Aligned: 34
Modelled: 34
Confidence: 43.6%
Identity: 24%
PDB header:transport protein
Chain: A: PDB Molecule:protein (band 3 anion transport protein);
PDBTitle: structural studies on the effects of the deletion in the2 red cell anion exchanger (band3, ae1) associated with3 south east asian ovalocytosis.
Phyre2
| 3 |
|
PDB 1pw4 chain A
Region: 12 - 463
Aligned: 413
Modelled: 429
Confidence: 37.6%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 4 |
|
PDB 1prt chain A
Region: 19 - 31
Aligned: 13
Modelled: 13
Confidence: 14.8%
Identity: 38%
Fold: ADP-ribosylation
Superfamily: ADP-ribosylation
Family: ADP-ribosylating toxins
Phyre2
| 5 |
|
PDB 1oyi chain A
Region: 18 - 31
Aligned: 14
Modelled: 14
Confidence: 14.3%
Identity: 43%
PDB header:viral protein
Chain: A: PDB Molecule:double-stranded rna-binding protein;
PDBTitle: solution structure of the z-dna binding domain of the2 vaccinia virus gene e3l
Phyre2
| 6 |
|
PDB 1oyi chain A
Region: 18 - 31
Aligned: 14
Modelled: 14
Confidence: 14.3%
Identity: 43%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Z-DNA binding domain
Phyre2
| 7 |
|
PDB 3lpz chain A
Region: 317 - 330
Aligned: 14
Modelled: 14
Confidence: 12.6%
Identity: 14%
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
Phyre2
| 8 |
|
PDB 3o7p chain A
Region: 36 - 462
Aligned: 403
Modelled: 427
Confidence: 6.1%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 9 |
|
PDB 1xmk chain A domain 1
Region: 18 - 33
Aligned: 16
Modelled: 16
Confidence: 5.6%
Identity: 25%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Z-DNA binding domain
Phyre2
| 10 |
|
PDB 2ht2 chain B
Region: 284 - 454
Aligned: 171
Modelled: 171
Confidence: 5.4%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 11 |
|
PDB 1zt2 chain A domain 1
Region: 2 - 27
Aligned: 26
Modelled: 26
Confidence: 5.2%
Identity: 15%
Fold: Prim-pol domain
Superfamily: Prim-pol domain
Family: PriA-like
Phyre2