| 1 |
|
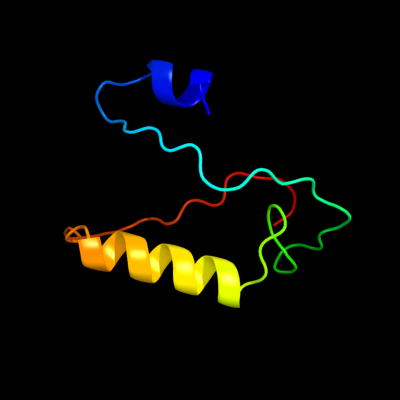
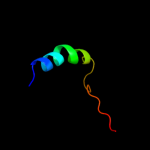
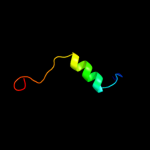
PDB 3pa8 chain A
Region: 9 - 72
Aligned: 64
Modelled: 64
Confidence: 25.7%
Identity: 20%
PDB header:toxin/peptide inhibitor
Chain: A: PDB Molecule:toxin b;
PDBTitle: structure of the c. difficile tcdb cysteine protease domain in complex2 with a peptide inhibitor
Phyre2
| 2 |
|
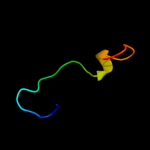
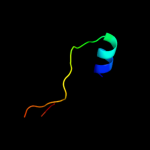
PDB 2zjr chain K domain 1
Region: 41 - 68
Aligned: 28
Modelled: 28
Confidence: 16.6%
Identity: 18%
Fold: Prokaryotic ribosomal protein L17
Superfamily: Prokaryotic ribosomal protein L17
Family: Prokaryotic ribosomal protein L17
Phyre2
| 3 |
|
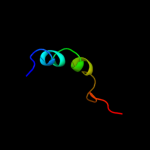
PDB 2cqm chain A domain 1
Region: 42 - 68
Aligned: 27
Modelled: 27
Confidence: 14.6%
Identity: 41%
Fold: Prokaryotic ribosomal protein L17
Superfamily: Prokaryotic ribosomal protein L17
Family: Prokaryotic ribosomal protein L17
Phyre2
| 4 |
|
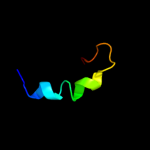
PDB 1tif chain A
Region: 1 - 31
Aligned: 30
Modelled: 31
Confidence: 12.8%
Identity: 33%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Translation initiation factor IF3, N-terminal domain
Family: Translation initiation factor IF3, N-terminal domain
Phyre2
| 5 |
|
PDB 1bnl chain A
Region: 21 - 39
Aligned: 19
Modelled: 19
Confidence: 10.6%
Identity: 32%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Endostatin
Phyre2
| 6 |
|
PDB 1gd8 chain A
Region: 41 - 70
Aligned: 30
Modelled: 30
Confidence: 9.6%
Identity: 23%
Fold: Prokaryotic ribosomal protein L17
Superfamily: Prokaryotic ribosomal protein L17
Family: Prokaryotic ribosomal protein L17
Phyre2
| 7 |
|
PDB 2jya chain A
Region: 31 - 57
Aligned: 27
Modelled: 27
Confidence: 9.0%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1810;
PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
Phyre2
| 8 |
|
PDB 1m9s chain A domain 3
Region: 23 - 44
Aligned: 22
Modelled: 22
Confidence: 8.2%
Identity: 18%
Fold: SH3-like barrel
Superfamily: Prokaryotic SH3-related domain
Family: GW domain
Phyre2
| 9 |
|
PDB 3bbo chain P
Region: 41 - 68
Aligned: 28
Modelled: 28
Confidence: 8.2%
Identity: 25%
PDB header:ribosome
Chain: P: PDB Molecule:ribosomal protein l17;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
Phyre2
| 10 |
|
PDB 1koe chain A
Region: 23 - 39
Aligned: 17
Modelled: 17
Confidence: 8.1%
Identity: 35%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Endostatin
Phyre2
| 11 |
|
PDB 2qam chain N domain 1
Region: 41 - 68
Aligned: 28
Modelled: 28
Confidence: 7.3%
Identity: 18%
Fold: Prokaryotic ribosomal protein L17
Superfamily: Prokaryotic ribosomal protein L17
Family: Prokaryotic ribosomal protein L17
Phyre2
| 12 |
|
PDB 1hd2 chain A
Region: 25 - 51
Aligned: 27
Modelled: 27
Confidence: 7.0%
Identity: 15%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione peroxidase-like
Phyre2
| 13 |
|
PDB 1ztx chain E domain 1
Region: 22 - 37
Aligned: 16
Modelled: 16
Confidence: 6.6%
Identity: 13%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: Class II viral fusion proteins C-terminal domain
Phyre2
| 14 |
|
PDB 2bh1 chain A domain 1
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 6.1%
Identity: 21%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Cyto-EpsL domain
Phyre2
| 15 |
|
PDB 2q79 chain A domain 1
Region: 11 - 35
Aligned: 22
Modelled: 25
Confidence: 6.1%
Identity: 27%
Fold: Ferredoxin-like
Superfamily: Viral DNA-binding domain
Family: Viral DNA-binding domain
Phyre2
| 16 |
|
PDB 2j01 chain S domain 1
Region: 24 - 38
Aligned: 15
Modelled: 15
Confidence: 5.9%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Translational machinery components
Family: Ribosomal protein L18 and S11
Phyre2
| 17 |
|
PDB 2jz8 chain A
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 33%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bh09830;
PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
Phyre2