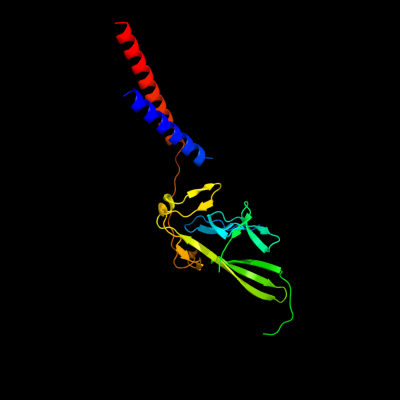
1 c3a69A_
100.0
51
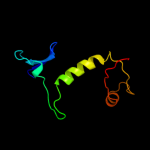
PDB header: motor proteinChain: A: PDB Molecule: flagellar hook protein flge;PDBTitle: atomic model of the bacterial flagellar hook based on2 docking an x-ray derived structure and terminal two alpha-3 helices into an 7.1 angstrom resolution cryoem map
2 d1wlga_
100.0
46
Fold: Flagellar hook protein flgESuperfamily: Flagellar hook protein flgEFamily: Flagellar hook protein flgE3 d1ucua_
85.1
21
Fold: Phase 1 flagellinSuperfamily: Phase 1 flagellinFamily: Phase 1 flagellin4 c1jzdA_
49.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: dsbc-dsbdalpha complex
5 c1oryB_
35.5
21
PDB header: chaperoneChain: B: PDB Molecule: flagellin;PDBTitle: flagellar export chaperone in complex with its cognate binding partner
6 c1zxuA_
28.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: at5g01750 protein;PDBTitle: x-ray structure of protein from arabidopsis thaliana2 at5g01750
7 d2q4ma1
28.2
17
Fold: Tubby C-terminal domain-likeSuperfamily: Tubby C-terminal domain-likeFamily: At5g01750-like8 c1t3bA_
28.1
15
PDB header: isomeraseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: x-ray structure of dsbc from haemophilus influenzae
9 c3k8vB_
27.6
18
PDB header: structural proteinChain: B: PDB Molecule: flagellin homolog;PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
10 d1niga_
17.9
24
Fold: Ferritin-likeSuperfamily: Cobalamin adenosyltransferase-likeFamily: Hypothetical protein Ta123811 d1lvoa_
15.8
36
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold12 c2q6fB_
15.2
36
PDB header: hydrolaseChain: B: PDB Molecule: infectious bronchitis virus (ibv) main protease;PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
13 c3d23A_
12.6
27
PDB header: hydrolaseChain: A: PDB Molecule: 3c-like proteinase;PDBTitle: main protease of hcov-hku1
14 d2duca1
12.6
27
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold15 c3kglB_
10.6
20
PDB header: plant proteinChain: B: PDB Molecule: cruciferin;PDBTitle: crystal structure of procruciferin, 11s globulin from2 brassica napus
16 d1t3ba2
10.1
15
Fold: Cystatin-likeSuperfamily: DsbC/DsbG N-terminal domain-likeFamily: DsbC/DsbG N-terminal domain-like17 d1p9sa_
10.1
18
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold18 d2v4jc1
9.8
42
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductaseSuperfamily: DsrC, the gamma subunit of dissimilatory sulfite reductaseFamily: DsrC, the gamma subunit of dissimilatory sulfite reductase19 c2oqkA_
9.3
18
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
20 d1eeja2
8.3
23
Fold: Cystatin-likeSuperfamily: DsbC/DsbG N-terminal domain-likeFamily: DsbC/DsbG N-terminal domain-like21 d1ji8a_
not modelled
7.9
21
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductaseSuperfamily: DsrC, the gamma subunit of dissimilatory sulfite reductaseFamily: DsrC, the gamma subunit of dissimilatory sulfite reductase22 d1b2pa_
not modelled
7.8
12
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins23 c2iyjA_
not modelled
7.8
23
PDB header: isomeraseChain: A: PDB Molecule: thiol disulfide interchange protein dsbc;PDBTitle: crystal structure of the n-terminal dimer domain of e.coli2 dsbc
24 c2dgyA_
not modelled
7.8
14
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
25 d2czra1
not modelled
7.4
32
Fold: Restriction endonuclease-likeSuperfamily: TBP-interacting protein-likeFamily: TBP-interacting protein-like26 c3bk2A_
not modelled
7.2
26
PDB header: hydrolaseChain: A: PDB Molecule: metal dependent hydrolase;PDBTitle: crystal structure analysis of the rnase j/ump complex
27 c2cazF_
not modelled
6.2
9
PDB header: protein transportChain: F: PDB Molecule: protein srn2;PDBTitle: escrt-i core
28 c1fxzC_
not modelled
6.0
18
PDB header: plant proteinChain: C: PDB Molecule: glycinin g1;PDBTitle: crystal structure of soybean proglycinin a1ab1b homotrimer
29 c2a5wC_
not modelled
6.0
19
PDB header: oxidoreductaseChain: C: PDB Molecule: sulfite reductase, desulfoviridin-type subunit gammaPDBTitle: crystal structure of the oxidized gamma-subunit of the dissimilatory2 sulfite reductase (dsrc) from archaeoglobus fulgidus
30 c3kscD_
not modelled
5.9
20
PDB header: plant proteinChain: D: PDB Molecule: lega class;PDBTitle: crystal structure of pea prolegumin, an 11s seed globulin2 from pisum sativum l.
31 d2enga_
not modelled
5.9
35
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: Eng V-like32 d2c5ra1
not modelled
5.5
33
Fold: Phage replication organizer domainSuperfamily: Phage replication organizer domainFamily: Phage replication organizer domain33 d1fxza2
not modelled
5.5
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein34 c1yx3A_
not modelled
5.3
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dsrc;PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
35 c2kjzA_
not modelled
5.2
29
PDB header: unknown functionChain: A: PDB Molecule: atc0852;PDBTitle: solution nmr structure of protein atc0852 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att2.