| 1 |
|
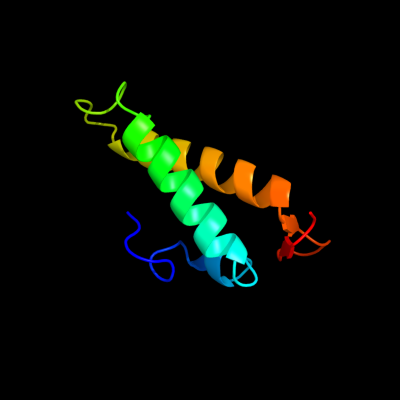
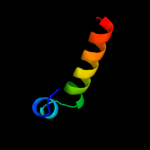
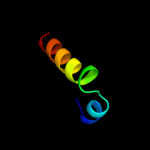
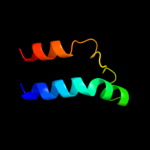
PDB 1sg7 chain A
Region: 2 - 76
Aligned: 75
Modelled: 75
Confidence: 100.0%
Identity: 100%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cation transport regulator chab;
PDBTitle: nmr solution structure of the putative cation transport2 regulator chab
Phyre2
| 2 |
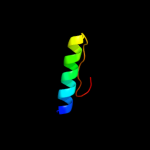
|
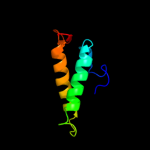
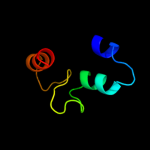
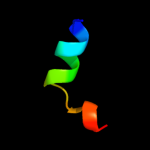
PDB 1sg7 chain A domain 1
Region: 2 - 76
Aligned: 75
Modelled: 75
Confidence: 100.0%
Identity: 100%
Fold: ChaB-like
Superfamily: ChaB-like
Family: ChaB-like
Phyre2
| 3 |
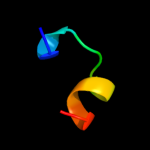
|
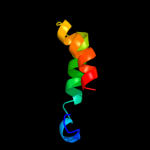
PDB 1k32 chain A domain 4
Region: 15 - 66
Aligned: 41
Modelled: 52
Confidence: 30.6%
Identity: 22%
Fold: ClpP/crotonase
Superfamily: ClpP/crotonase
Family: Tail specific protease, catalytic domain
Phyre2
| 4 |
|
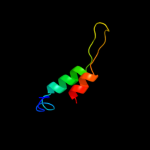
PDB 1fc6 chain A domain 4
Region: 22 - 66
Aligned: 34
Modelled: 45
Confidence: 28.1%
Identity: 32%
Fold: ClpP/crotonase
Superfamily: ClpP/crotonase
Family: Tail specific protease, catalytic domain
Phyre2
| 5 |
|
PDB 2q9q chain C
Region: 9 - 37
Aligned: 28
Modelled: 29
Confidence: 22.7%
Identity: 11%
PDB header:replication
Chain: C: PDB Molecule:dna replication complex gins protein psf1;
PDBTitle: the crystal structure of full length human gins complex
Phyre2
| 6 |
|
PDB 1knc chain A
Region: 11 - 59
Aligned: 39
Modelled: 49
Confidence: 18.5%
Identity: 26%
Fold: AhpD-like
Superfamily: AhpD-like
Family: AhpD
Phyre2
| 7 |
|
PDB 1bpo chain A
Region: 13 - 56
Aligned: 32
Modelled: 44
Confidence: 17.5%
Identity: 31%
PDB header:membrane protein
Chain: A: PDB Molecule:protein (clathrin);
PDBTitle: clathrin heavy-chain terminal domain and linker
Phyre2
| 8 |
|
PDB 1bpo chain A domain 1
Region: 12 - 56
Aligned: 34
Modelled: 45
Confidence: 16.4%
Identity: 26%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: Clathrin heavy-chain linker domain
Phyre2
| 9 |
|
PDB 2e9x chain A domain 1
Region: 9 - 36
Aligned: 27
Modelled: 28
Confidence: 15.8%
Identity: 11%
Fold: GINS helical bundle-like
Superfamily: GINS helical bundle-like
Family: PSF1 N-terminal domain-like
Phyre2
| 10 |
|
PDB 3mop chain M
Region: 6 - 17
Aligned: 12
Modelled: 12
Confidence: 11.0%
Identity: 33%
PDB header:signaling protein, immune system
Chain: M: PDB Molecule:interleukin-1 receptor-associated kinase-like 2;
PDBTitle: the ternary death domain complex of myd88, irak4, and irak2
Phyre2
| 11 |
|
PDB 1o8t chain A
Region: 21 - 35
Aligned: 15
Modelled: 15
Confidence: 8.8%
Identity: 33%
PDB header:lipid transport
Chain: A: PDB Molecule:apolipoprotein c-ii;
PDBTitle: global structure and dynamics of human apolipoprotein cii2 in complex with micelles: evidence for increased mobility3 of the helix involvved in the activation of lipoprotein4 lipase.
Phyre2
| 12 |
|
PDB 1trl chain B
Region: 44 - 60
Aligned: 17
Modelled: 17
Confidence: 8.2%
Identity: 35%
PDB header:hydrolase (metalloprotease)
Chain: B: PDB Molecule:thermolysin fragment 255 - 316;
PDBTitle: nmr solution structure of the c-terminal fragment 255-3162 of thermolysin: a dimer formed by subunits having the3 native structure
Phyre2
| 13 |
|
PDB 2qfk chain A domain 1
Region: 11 - 31
Aligned: 21
Modelled: 21
Confidence: 6.4%
Identity: 29%
Fold: Globin-like
Superfamily: Globin-like
Family: Globins
Phyre2
| 14 |
|
PDB 1fc9 chain A
Region: 22 - 66
Aligned: 34
Modelled: 45
Confidence: 5.7%
Identity: 32%
PDB header:hydrolase
Chain: A: PDB Molecule:photosystem ii d1 protease;
PDBTitle: photosystem ii d1 c-terminal processing protease
Phyre2
| 15 |
|
PDB 2ddz chain A domain 1
Region: 19 - 47
Aligned: 29
Modelled: 29
Confidence: 5.7%
Identity: 21%
Fold: Class II aaRS and biotin synthetases
Superfamily: Class II aaRS and biotin synthetases
Family: PH0223-like
Phyre2
| 16 |
|
PDB 2kwu chain A
Region: 7 - 17
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 45%
PDB header:protein binding/signaling protein
Chain: A: PDB Molecule:dna polymerase iota;
PDBTitle: solution structure of ubm2 of murine polymerase iota in complex with2 ubiquitin
Phyre2
| 17 |
|
PDB 1tns chain A
Region: 6 - 17
Aligned: 12
Modelled: 12
Confidence: 5.3%
Identity: 33%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Excisionase-like
Phyre2
| 18 |
|
PDB 1qpm chain A
Region: 5 - 17
Aligned: 13
Modelled: 13
Confidence: 5.2%
Identity: 8%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Excisionase-like
Phyre2