| 1 |
|
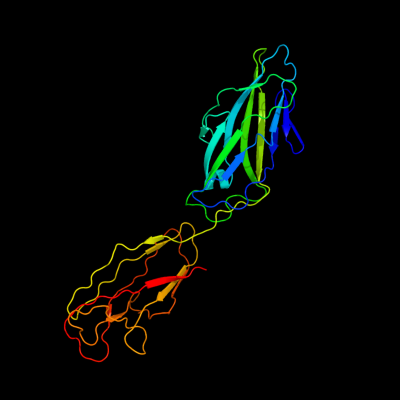
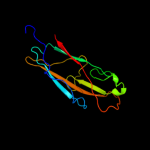
PDB 1klf chain P
Region: 22 - 300
Aligned: 279
Modelled: 279
Confidence: 100.0%
Identity: 100%
PDB header:chaperone/adhesin complex
Chain: P: PDB Molecule:fimh protein;
PDBTitle: fimh adhesin-fimc chaperone complex with d-mannose
Phyre2
| 2 |
|
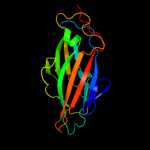
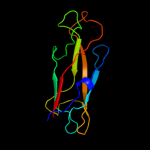
PDB 1ze3 chain H domain 1
Region: 180 - 300
Aligned: 121
Modelled: 121
Confidence: 100.0%
Identity: 100%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 3 |
|
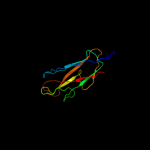
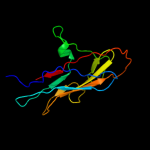
PDB 1uwf chain A domain 1
Region: 22 - 179
Aligned: 158
Modelled: 158
Confidence: 99.9%
Identity: 100%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 4 |
|
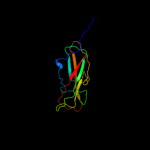
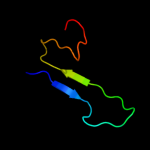
PDB 3bwu chain F
Region: 181 - 300
Aligned: 119
Modelled: 120
Confidence: 99.9%
Identity: 24%
PDB header:chaperone, structural, membrane protein
Chain: F: PDB Molecule:protein fimf;
PDBTitle: crystal structure of the ternary complex of fimd (n-terminal domain,2 fimdn) with fimc and the n-terminally truncated pilus subunit fimf3 (fimft)
Phyre2
| 5 |
|
PDB 3jwn chain K
Region: 169 - 300
Aligned: 132
Modelled: 132
Confidence: 99.9%
Identity: 23%
PDB header:protein binding/cell adhesion
Chain: K: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 6 |
|
PDB 3jwn chain L
Region: 169 - 300
Aligned: 132
Modelled: 132
Confidence: 99.9%
Identity: 23%
PDB header:protein binding/cell adhesion
Chain: L: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 7 |
|
PDB 3jwn chain F
Region: 169 - 300
Aligned: 132
Modelled: 132
Confidence: 99.9%
Identity: 23%
PDB header:protein binding/cell adhesion
Chain: F: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 8 |
|
PDB 3jwn chain E
Region: 169 - 300
Aligned: 132
Modelled: 132
Confidence: 99.9%
Identity: 23%
PDB header:protein binding/cell adhesion
Chain: E: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 9 |
|
PDB 2jmr chain A
Region: 169 - 300
Aligned: 132
Modelled: 132
Confidence: 99.9%
Identity: 20%
PDB header:cell adhesion
Chain: A: PDB Molecule:fimf;
PDBTitle: nmr structure of the e. coli type 1 pilus subunit fimf
Phyre2
| 10 |
|
PDB 3bfw chain A
Region: 179 - 299
Aligned: 118
Modelled: 121
Confidence: 99.9%
Identity: 20%
PDB header:structural protein/structural protein
Chain: A: PDB Molecule:protein fimg;
PDBTitle: crystal structure of truncated fimg (fimgt) in complex with the donor2 strand peptide of fimf (dsf)
Phyre2
| 11 |
|
PDB 2j2z chain B domain 1
Region: 169 - 300
Aligned: 128
Modelled: 132
Confidence: 99.8%
Identity: 18%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 12 |
|
PDB 2jty chain A
Region: 169 - 300
Aligned: 131
Modelled: 132
Confidence: 99.8%
Identity: 24%
PDB header:structural protein
Chain: A: PDB Molecule:type-1 fimbrial protein, a chain;
PDBTitle: self-complemented variant of fima, the main subunit of type 1 pilus
Phyre2
| 13 |
|
PDB 1pdk chain B
Region: 177 - 300
Aligned: 122
Modelled: 124
Confidence: 99.8%
Identity: 18%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 14 |
|
PDB 2uy6 chain B domain 1
Region: 169 - 300
Aligned: 128
Modelled: 132
Confidence: 99.8%
Identity: 13%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 15 |
|
PDB 2w07 chain B
Region: 176 - 300
Aligned: 110
Modelled: 125
Confidence: 99.7%
Identity: 20%
PDB header:cell adhesion
Chain: B: PDB Molecule:minor pilin subunit papf;
PDBTitle: structural determinants of polymerization reactivity of the2 p pilus adaptor subunit papf
Phyre2
| 16 |
|
PDB 1n12 chain A
Region: 180 - 299
Aligned: 118
Modelled: 120
Confidence: 99.4%
Identity: 17%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 17 |
|
PDB 2wmp chain B
Region: 179 - 299
Aligned: 110
Modelled: 121
Confidence: 91.2%
Identity: 18%
PDB header:chaperone
Chain: B: PDB Molecule:papg protein;
PDBTitle: structure of the e. coli chaperone papd in complex with the pilin2 domain of the papgii adhesin
Phyre2
| 18 |
|
PDB 2jna chain A domain 1
Region: 1 - 22
Aligned: 22
Modelled: 22
Confidence: 32.8%
Identity: 27%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 19 |
|
PDB 2noc chain A domain 1
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 13.9%
Identity: 17%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 20 |
|
PDB 2oz4 chain A domain 1
Region: 187 - 225
Aligned: 39
Modelled: 39
Confidence: 6.8%
Identity: 23%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: C2 set domains
Phyre2
| 21 |
|
| 22 |
|