| 1 |
|
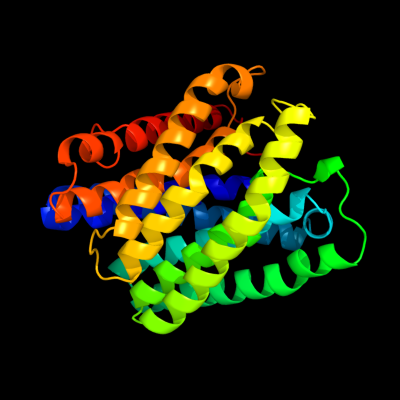
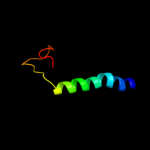
PDB 3m7b chain A
Region: 9 - 316
Aligned: 304
Modelled: 305
Confidence: 100.0%
Identity: 40%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 2 |
|
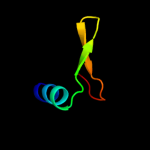
PDB 2knc chain A
Region: 275 - 327
Aligned: 50
Modelled: 53
Confidence: 12.1%
Identity: 10%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 3 |
|
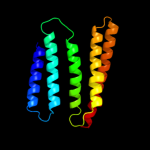
PDB 3kys chain B
Region: 5 - 14
Aligned: 10
Modelled: 10
Confidence: 8.9%
Identity: 30%
PDB header:transcription/protein binding
Chain: B: PDB Molecule:65 kda yes-associated protein;
PDBTitle: crystal structure of human yap and tead complex
Phyre2
| 4 |
|
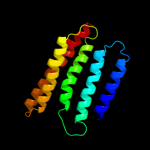
PDB 1cn3 chain F
Region: 135 - 146
Aligned: 12
Modelled: 12
Confidence: 8.3%
Identity: 33%
PDB header:viral protein
Chain: F: PDB Molecule:fragment of coat protein vp2;
PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
Phyre2
| 5 |
|
PDB 3dww chain A
Region: 288 - 330
Aligned: 43
Modelled: 43
Confidence: 7.0%
Identity: 12%
PDB header:isomerase
Chain: A: PDB Molecule:prostaglandin e synthase;
PDBTitle: electron crystallographic structure of human microsomal2 prostaglandin e synthase 1
Phyre2
| 6 |
|
PDB 3kcq chain A
Region: 301 - 330
Aligned: 30
Modelled: 30
Confidence: 6.3%
Identity: 20%
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
Phyre2
| 7 |
|
PDB 1i25 chain A
Region: 322 - 329
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 38%
PDB header:toxin
Chain: A: PDB Molecule:huwentoxin-ii;
PDBTitle: three dimensional solution structure of huwentoxin-ii by 2d2 1h-nmr
Phyre2
| 8 |
|
PDB 1i25 chain A
Region: 322 - 329
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 38%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: omega toxin-like
Family: Spider toxins
Phyre2
| 9 |
|
PDB 1xio chain A
Region: 173 - 313
Aligned: 140
Modelled: 141
Confidence: 5.8%
Identity: 8%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 10 |
|
PDB 1xio chain A
Region: 173 - 313
Aligned: 140
Modelled: 141
Confidence: 5.8%
Identity: 8%
PDB header:signaling protein
Chain: A: PDB Molecule:anabaena sensory rhodopsin;
PDBTitle: anabaena sensory rhodopsin
Phyre2
| 11 |
|
PDB 4cro chain A
Region: 311 - 330
Aligned: 20
Modelled: 20
Confidence: 5.5%
Identity: 15%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: Phage repressors
Phyre2
| 12 |
|
PDB 2rm9 chain A
Region: 322 - 330
Aligned: 9
Modelled: 9
Confidence: 5.4%
Identity: 22%
PDB header:neuropeptide
Chain: A: PDB Molecule:astressin2b;
PDBTitle: astressin2b
Phyre2