| 1 |
|
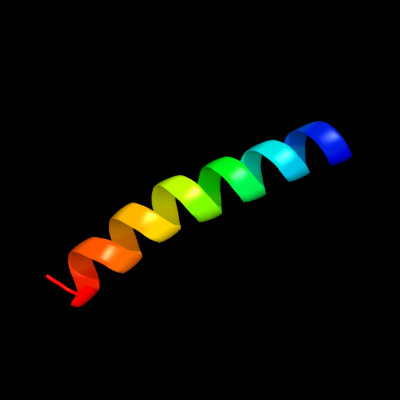
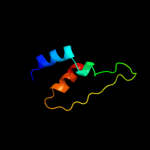
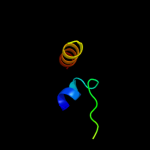
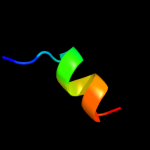
PDB 2dw3 chain A
Region: 78 - 104
Aligned: 27
Modelled: 27
Confidence: 54.1%
Identity: 30%
PDB header:photosynthesis
Chain: A: PDB Molecule:intrinsic membrane protein pufx;
PDBTitle: solution structure of the rhodobacter sphaeroides pufx2 membrane protein
Phyre2
| 2 |
|
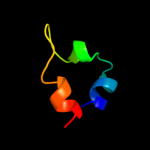
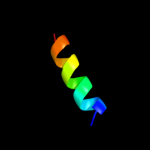
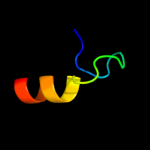
PDB 1ysg chain A domain 1
Region: 89 - 113
Aligned: 17
Modelled: 25
Confidence: 19.0%
Identity: 29%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 3 |
|
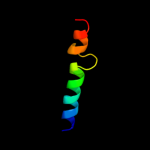
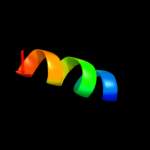
PDB 1g5m chain A
Region: 89 - 113
Aligned: 17
Modelled: 25
Confidence: 16.3%
Identity: 24%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 4 |
|
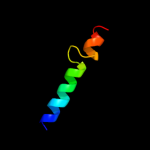
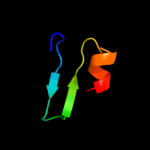
PDB 2pon chain B domain 1
Region: 89 - 113
Aligned: 17
Modelled: 25
Confidence: 14.4%
Identity: 29%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 5 |
|
PDB 1kl7 chain A
Region: 90 - 139
Aligned: 47
Modelled: 50
Confidence: 12.7%
Identity: 26%
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family: Tryptophan synthase beta subunit-like PLP-dependent enzymes
Phyre2
| 6 |
|
PDB 1s4k chain A
Region: 38 - 66
Aligned: 29
Modelled: 29
Confidence: 12.5%
Identity: 17%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: YdiL-like
Phyre2
| 7 |
|
PDB 1ifl chain A
Region: 20 - 32
Aligned: 13
Modelled: 13
Confidence: 11.2%
Identity: 46%
PDB header:virus
Chain: A: PDB Molecule:inovirus;
PDBTitle: molecular models and structural comparisons of native and2 mutant class i filamentous bacteriophages ff (fd, f1, m13),3 if1 and ike
Phyre2
| 8 |
|
PDB 2k9y chain B
Region: 30 - 58
Aligned: 26
Modelled: 29
Confidence: 10.3%
Identity: 38%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 9 |
|
PDB 2k9y chain A
Region: 30 - 58
Aligned: 26
Modelled: 29
Confidence: 10.3%
Identity: 38%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 10 |
|
PDB 2wvr chain C
Region: 110 - 139
Aligned: 30
Modelled: 30
Confidence: 9.7%
Identity: 20%
PDB header:replication
Chain: C: PDB Molecule:dna replication factor cdt1;
PDBTitle: human cdt1:geminin complex
Phyre2
| 11 |
|
PDB 2bol chain A
Region: 49 - 64
Aligned: 16
Modelled: 16
Confidence: 9.0%
Identity: 19%
PDB header:heat shock protein
Chain: A: PDB Molecule:small heat shock protein;
PDBTitle: crystal structure and assembly of tsp36, a metazoan small2 heat shock protein
Phyre2
| 12 |
|
PDB 1i7q chain B
Region: 116 - 136
Aligned: 19
Modelled: 21
Confidence: 9.0%
Identity: 21%
Fold: Flavodoxin-like
Superfamily: Class I glutamine amidotransferase-like
Family: Class I glutamine amidotransferases (GAT)
Phyre2
| 13 |
|
PDB 1xrx chain A domain 1
Region: 125 - 136
Aligned: 12
Modelled: 10
Confidence: 8.6%
Identity: 33%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: SeqA N-terminal domain-like
Phyre2
| 14 |
|
PDB 1xrx chain D
Region: 125 - 136
Aligned: 12
Modelled: 10
Confidence: 8.6%
Identity: 33%
PDB header:replication inhibitor
Chain: D: PDB Molecule:seqa protein;
PDBTitle: crystal structure of a dna-binding protein
Phyre2
| 15 |
|
PDB 2p7t chain C domain 1
Region: 16 - 30
Aligned: 14
Modelled: 15
Confidence: 8.4%
Identity: 36%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 16 |
|
PDB 1wlo chain A
Region: 110 - 136
Aligned: 27
Modelled: 27
Confidence: 8.2%
Identity: 4%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:sufe protein;
PDBTitle: solution structure of the hypothetical protein from thermus2 thermophilus hb8
Phyre2
| 17 |
|
PDB 2j6b chain A domain 1
Region: 123 - 141
Aligned: 19
Modelled: 19
Confidence: 7.1%
Identity: 21%
Fold: STIV B116-like
Superfamily: STIV B116-like
Family: STIV B116-like
Phyre2
| 18 |
|
PDB 2x4i chain A
Region: 122 - 141
Aligned: 20
Modelled: 20
Confidence: 6.6%
Identity: 25%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein 114;
PDBTitle: orf 114a from sulfolobus islandicus rudivirus 1
Phyre2
| 19 |
|
PDB 3fmt chain F
Region: 125 - 136
Aligned: 12
Modelled: 12
Confidence: 6.1%
Identity: 33%
PDB header:replication inhibitor/dna
Chain: F: PDB Molecule:protein seqa;
PDBTitle: crystal structure of seqa bound to dna
Phyre2
| 20 |
|
PDB 1i1q chain B
Region: 116 - 136
Aligned: 19
Modelled: 21
Confidence: 6.0%
Identity: 21%
Fold: Flavodoxin-like
Superfamily: Class I glutamine amidotransferase-like
Family: Class I glutamine amidotransferases (GAT)
Phyre2
| 21 |
|
| 22 |
|