| 1 |
|
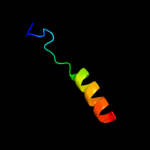
PDB 2knc chain A
Region: 69 - 92
Aligned: 24
Modelled: 24
Confidence: 35.6%
Identity: 29%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 2 |
|
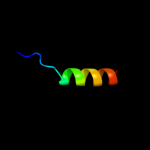
PDB 2k1a chain A
Region: 73 - 92
Aligned: 20
Modelled: 20
Confidence: 23.4%
Identity: 30%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 3 |
|
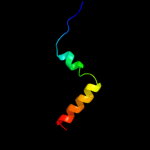
PDB 2voy chain B
Region: 12 - 45
Aligned: 34
Modelled: 34
Confidence: 22.0%
Identity: 18%
PDB header:hydrolase
Chain: B: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 4 |
|
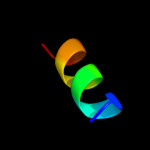
PDB 2jtw chain A
Region: 16 - 36
Aligned: 21
Modelled: 21
Confidence: 7.3%
Identity: 43%
PDB header:membrane protein
Chain: A: PDB Molecule:transmembrane helix 7 of yeast vatpase;
PDBTitle: solution structure of tm7 bound to dpc micelles
Phyre2
| 5 |
|
PDB 2yt9 chain A domain 2
Region: 20 - 32
Aligned: 13
Modelled: 13
Confidence: 6.1%
Identity: 31%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 6 |
|
PDB 3dl8 chain F
Region: 59 - 76
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 33%
PDB header:protein transport
Chain: F: PDB Molecule:protein-export membrane protein secg;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
Phyre2
| 7 |
|
PDB 3da5 chain A
Region: 12 - 23
Aligned: 12
Modelled: 12
Confidence: 6.1%
Identity: 50%
PDB header:rna binding protein
Chain: A: PDB Molecule:argonaute;
PDBTitle: crystal structure of piwi/argonaute/zwille(paz) domain from2 thermococcus thioreducens
Phyre2