| 1 |
|
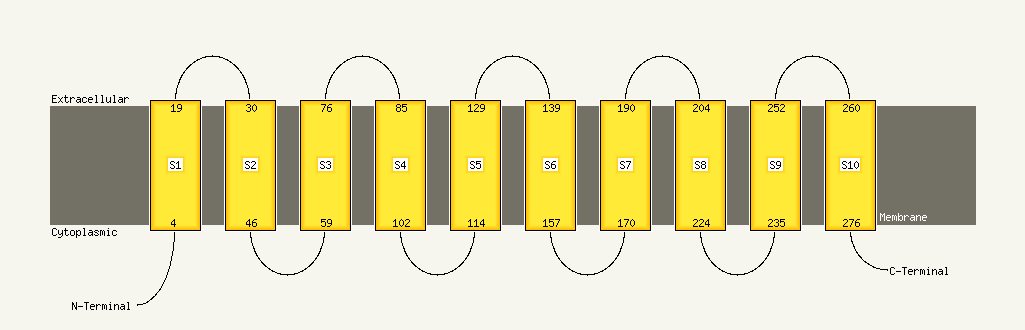
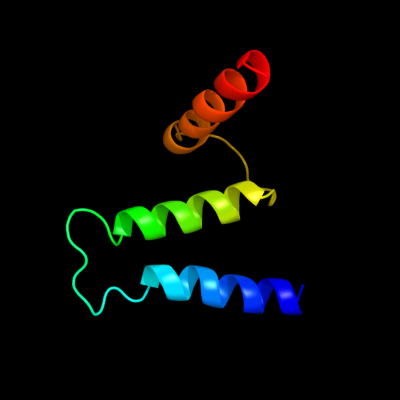
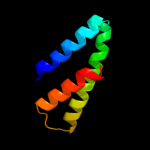
PDB 1s7b chain A
Region: 210 - 276
Aligned: 67
Modelled: 67
Confidence: 97.3%
Identity: 10%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 2 |
|
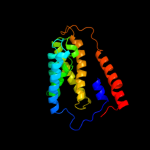
PDB 2i68 chain B
Region: 214 - 276
Aligned: 57
Modelled: 63
Confidence: 42.8%
Identity: 16%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 3 |
|
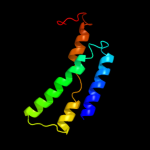
PDB 1ifp chain A
Region: 263 - 281
Aligned: 19
Modelled: 0
Confidence: 31.8%
Identity: 32%
PDB header:virus
Chain: A: PDB Molecule:major coat protein assembly;
PDBTitle: inovirus (filamentous bacteriophage) strain pf3 major coat2 protein assembly
Phyre2
| 4 |
|
PDB 2jx4 chain A
Region: 282 - 297
Aligned: 16
Modelled: 16
Confidence: 9.2%
Identity: 38%
PDB header:membrane protein
Chain: A: PDB Molecule:vasopressin v2 receptor;
PDBTitle: nmr structure of the intracellular loop (i3) of the2 vasopressin v2 receptor (gpcr)
Phyre2
| 5 |
|
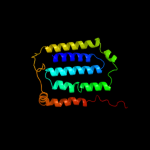
PDB 3aqp chain B
Region: 208 - 273
Aligned: 66
Modelled: 66
Confidence: 8.8%
Identity: 12%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 6 |
|
PDB 1pw4 chain A
Region: 114 - 290
Aligned: 177
Modelled: 177
Confidence: 8.5%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 7 |
|
PDB 1u7g chain A
Region: 37 - 234
Aligned: 194
Modelled: 198
Confidence: 8.0%
Identity: 12%
Fold: Ammonium transporter
Superfamily: Ammonium transporter
Family: Ammonium transporter
Phyre2
| 8 |
|
PDB 1iwg chain A domain 7
Region: 207 - 293
Aligned: 87
Modelled: 87
Confidence: 7.6%
Identity: 14%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 9 |
|
PDB 3rko chain F
Region: 118 - 284
Aligned: 163
Modelled: 167
Confidence: 7.2%
Identity: 10%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 261 - 294
Aligned: 34
Modelled: 34
Confidence: 5.8%
Identity: 18%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2