| 1 |
|
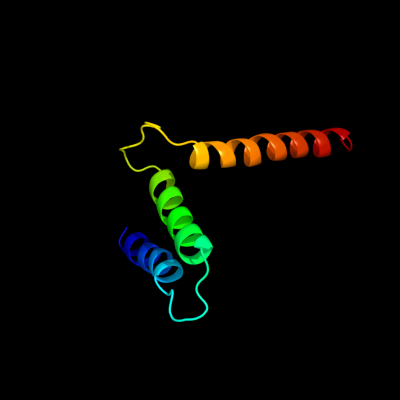
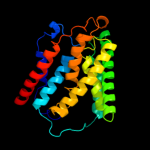
PDB 1s7b chain A
Region: 223 - 297
Aligned: 75
Modelled: 75
Confidence: 97.7%
Identity: 19%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 2 |
|
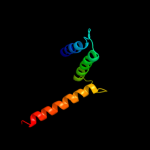
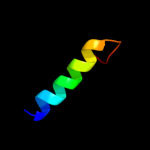
PDB 2i68 chain B
Region: 227 - 290
Aligned: 58
Modelled: 64
Confidence: 91.5%
Identity: 21%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 3 |
|
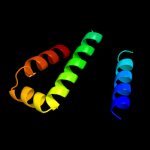
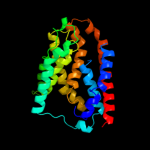
PDB 3rko chain F
Region: 159 - 298
Aligned: 138
Modelled: 140
Confidence: 68.7%
Identity: 17%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 4 |
|
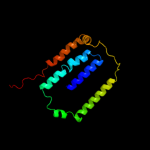
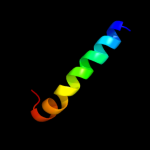
PDB 2b2h chain A
Region: 4 - 243
Aligned: 238
Modelled: 240
Confidence: 40.2%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 5 |
|
PDB 2l2t chain A
Region: 278 - 300
Aligned: 23
Modelled: 23
Confidence: 20.2%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 4 - 243
Aligned: 229
Modelled: 240
Confidence: 15.8%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 2jp3 chain A
Region: 275 - 300
Aligned: 26
Modelled: 26
Confidence: 10.7%
Identity: 19%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 8 |
|
PDB 1u7g chain A
Region: 11 - 243
Aligned: 232
Modelled: 233
Confidence: 9.6%
Identity: 8%
Fold: Ammonium transporter
Superfamily: Ammonium transporter
Family: Ammonium transporter
Phyre2
| 9 |
|
PDB 2jo1 chain A
Region: 275 - 300
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 12%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 10 |
|
PDB 3aqp chain B
Region: 214 - 300
Aligned: 87
Modelled: 87
Confidence: 5.7%
Identity: 17%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2