| 1 |
|
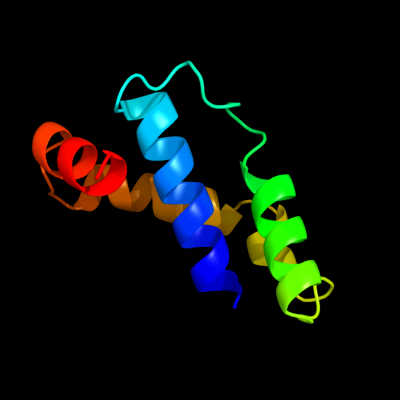
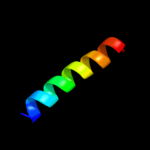
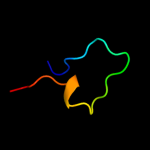
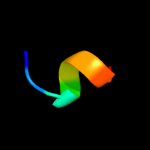
PDB 3utk chain A
Region: 27 - 105
Aligned: 77
Modelled: 79
Confidence: 95.5%
Identity: 31%
PDB header:protein transport
Chain: A: PDB Molecule:lipoprotein outs;
PDBTitle: structure of the pilotin of the type ii secretion system
Phyre2
| 2 |
|
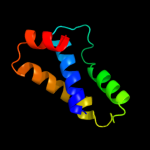
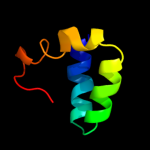
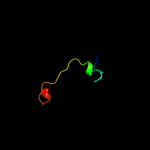
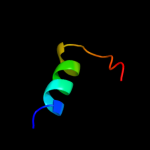
PDB 3sol chain A
Region: 27 - 105
Aligned: 77
Modelled: 79
Confidence: 93.1%
Identity: 25%
PDB header:protein transport
Chain: A: PDB Molecule:type ii secretion pathway related protein;
PDBTitle: crystal structure of the type 2 secretion system pilotin gsps
Phyre2
| 3 |
|
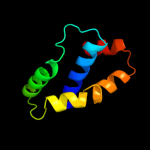
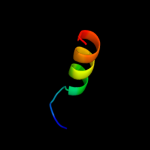
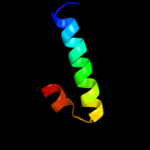
PDB 4a56 chain A
Region: 27 - 105
Aligned: 77
Modelled: 79
Confidence: 92.2%
Identity: 26%
PDB header:protein transport
Chain: A: PDB Molecule:pullulanase secretion protein puls;
PDBTitle: crystal structure of the type 2 secretion system pilotin2 from klebsiella oxytoca
Phyre2
| 4 |
|
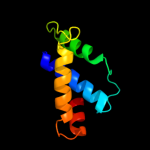
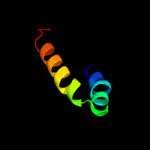
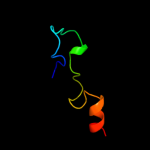
PDB 2al6 chain A domain 2
Region: 24 - 44
Aligned: 21
Modelled: 21
Confidence: 42.5%
Identity: 43%
Fold: PH domain-like barrel
Superfamily: PH domain-like
Family: Third domain of FERM
Phyre2
| 5 |
|
PDB 3pnt chain D
Region: 30 - 77
Aligned: 47
Modelled: 48
Confidence: 20.2%
Identity: 28%
PDB header:hydrolase/hydrolase inhibitor
Chain: D: PDB Molecule:immunity factor for spn;
PDBTitle: crystal structure of the streptococcus pyogenes nad+ glycohydrolase2 spn in complex with ifs, the immunity factor for spn
Phyre2
| 6 |
|
PDB 1q0q chain A domain 1
Region: 32 - 59
Aligned: 27
Modelled: 28
Confidence: 15.3%
Identity: 19%
Fold: Left-handed superhelix
Superfamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain
Family: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain
Phyre2
| 7 |
|
PDB 1wn4 chain A
Region: 48 - 64
Aligned: 17
Modelled: 17
Confidence: 14.1%
Identity: 41%
PDB header:plant protein
Chain: A: PDB Molecule:vontr protein;
PDBTitle: nmr structure of vontr
Phyre2
| 8 |
|
PDB 3qb2 chain A
Region: 30 - 67
Aligned: 37
Modelled: 38
Confidence: 13.5%
Identity: 24%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:immunity factor for spn;
PDBTitle: the crystal structure of immunity factor for spn (ifs)
Phyre2
| 9 |
|
PDB 1r0k chain A domain 1
Region: 32 - 52
Aligned: 21
Modelled: 21
Confidence: 11.6%
Identity: 29%
Fold: Left-handed superhelix
Superfamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain
Family: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain
Phyre2
| 10 |
|
PDB 1mi1 chain A
Region: 68 - 99
Aligned: 32
Modelled: 32
Confidence: 11.5%
Identity: 28%
PDB header:signaling protein
Chain: A: PDB Molecule:neurobeachin;
PDBTitle: crystal structure of the ph-beach domain of human2 neurobeachin
Phyre2
| 11 |
|
PDB 1t77 chain A domain 1
Region: 68 - 90
Aligned: 23
Modelled: 23
Confidence: 10.9%
Identity: 30%
Fold: BEACH domain
Superfamily: BEACH domain
Family: BEACH domain
Phyre2
| 12 |
|
PDB 3dal chain A
Region: 74 - 108
Aligned: 34
Modelled: 35
Confidence: 9.7%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:pr domain zinc finger protein 1;
PDBTitle: methyltransferase domain of human pr domain-containing2 protein 1
Phyre2
| 13 |
|
PDB 2i2x chain O
Region: 44 - 49
Aligned: 6
Modelled: 6
Confidence: 8.5%
Identity: 83%
PDB header:transferase
Chain: O: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
Phyre2
| 14 |
|
PDB 3frw chain F
Region: 23 - 53
Aligned: 31
Modelled: 31
Confidence: 8.0%
Identity: 13%
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative trp repressor protein;
PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
Phyre2
| 15 |
|
PDB 1mi1 chain A domain 1
Region: 68 - 106
Aligned: 39
Modelled: 39
Confidence: 7.9%
Identity: 23%
Fold: BEACH domain
Superfamily: BEACH domain
Family: BEACH domain
Phyre2
| 16 |
|
PDB 3a14 chain B
Region: 32 - 52
Aligned: 21
Modelled: 21
Confidence: 7.8%
Identity: 33%
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
Phyre2
| 17 |
|
PDB 3kor chain D
Region: 23 - 53
Aligned: 31
Modelled: 31
Confidence: 7.5%
Identity: 13%
PDB header:transcription
Chain: D: PDB Molecule:possible trp repressor;
PDBTitle: crystal structure of a putative trp repressor from staphylococcus2 aureus
Phyre2
| 18 |
|
PDB 2wx3 chain A
Region: 31 - 38
Aligned: 8
Modelled: 8
Confidence: 7.0%
Identity: 50%
PDB header:structural protein
Chain: A: PDB Molecule:mrna-decapping enzyme 1a;
PDBTitle: asymmetric trimer of the human dcp1a c-terminal domain
Phyre2
| 19 |
|
PDB 3mcr chain A
Region: 32 - 52
Aligned: 21
Modelled: 21
Confidence: 6.1%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh dehydrogenase, subunit c;
PDBTitle: crystal structure of nadh dehydrogenase subunit c (tfu_2693) from2 thermobifida fusca yx-er1 at 2.65 a resolution
Phyre2
| 20 |
|
PDB 2jcy chain A
Region: 32 - 52
Aligned: 21
Modelled: 21
Confidence: 6.0%
Identity: 24%
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium3 tuberculosis
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|