| 1 |
|
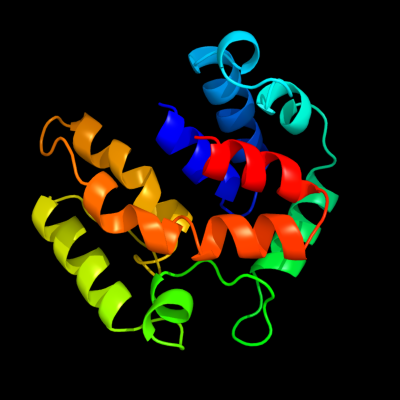
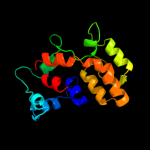
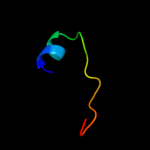
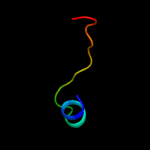
PDB 2o9x chain A domain 1
Region: 2 - 157
Aligned: 146
Modelled: 156
Confidence: 100.0%
Identity: 22%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 2 |
|
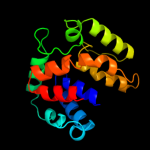
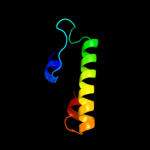
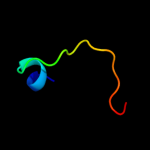
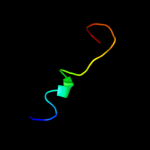
PDB 2o9x chain A
Region: 2 - 157
Aligned: 146
Modelled: 156
Confidence: 99.9%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:reductase, assembly protein;
PDBTitle: crystal structure of a putative redox enzyme maturation protein from2 archaeoglobus fulgidus
Phyre2
| 3 |
|
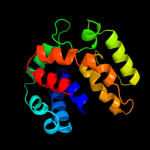
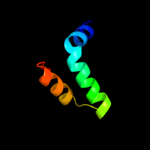
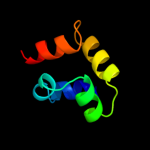
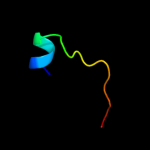
PDB 1n1c chain A
Region: 2 - 156
Aligned: 151
Modelled: 155
Confidence: 99.8%
Identity: 19%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 4 |
|
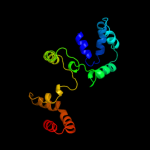
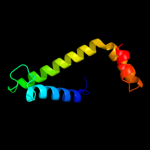
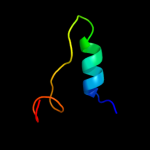
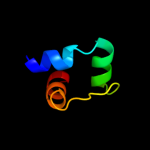
PDB 1s9u chain A
Region: 3 - 154
Aligned: 148
Modelled: 152
Confidence: 99.8%
Identity: 18%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 5 |
|
PDB 2idg chain A domain 1
Region: 87 - 139
Aligned: 52
Modelled: 52
Confidence: 98.5%
Identity: 17%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 6 |
|
PDB 2di4 chain B
Region: 120 - 163
Aligned: 44
Modelled: 44
Confidence: 16.1%
Identity: 20%
PDB header:hydrolase
Chain: B: PDB Molecule:cell division protein ftsh homolog;
PDBTitle: crystal structure of the ftsh protease domain
Phyre2
| 7 |
|
PDB 1lva chain A domain 3
Region: 77 - 100
Aligned: 24
Modelled: 24
Confidence: 13.1%
Identity: 21%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: C-terminal fragment of elongation factor SelB
Phyre2
| 8 |
|
PDB 2ce7 chain A domain 1
Region: 72 - 156
Aligned: 85
Modelled: 85
Confidence: 8.9%
Identity: 14%
Fold: FtsH protease domain-like
Superfamily: FtsH protease domain-like
Family: FtsH protease domain-like
Phyre2
| 9 |
|
PDB 1ghe chain A
Region: 88 - 106
Aligned: 18
Modelled: 19
Confidence: 8.8%
Identity: 33%
Fold: Acyl-CoA N-acyltransferases (Nat)
Superfamily: Acyl-CoA N-acyltransferases (Nat)
Family: N-acetyl transferase, NAT
Phyre2
| 10 |
|
PDB 2g3a chain A domain 1
Region: 88 - 106
Aligned: 18
Modelled: 19
Confidence: 8.7%
Identity: 28%
Fold: Acyl-CoA N-acyltransferases (Nat)
Superfamily: Acyl-CoA N-acyltransferases (Nat)
Family: N-acetyl transferase, NAT
Phyre2
| 11 |
|
PDB 1yx0 chain A domain 1
Region: 88 - 106
Aligned: 18
Modelled: 19
Confidence: 7.8%
Identity: 39%
Fold: Acyl-CoA N-acyltransferases (Nat)
Superfamily: Acyl-CoA N-acyltransferases (Nat)
Family: N-acetyl transferase, NAT
Phyre2
| 12 |
|
PDB 3iwf chain A
Region: 4 - 57
Aligned: 54
Modelled: 54
Confidence: 7.4%
Identity: 6%
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator rpir family;
PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
Phyre2
| 13 |
|
PDB 2d6f chain C domain 2
Region: 77 - 106
Aligned: 29
Modelled: 30
Confidence: 7.0%
Identity: 31%
Fold: DCoH-like
Superfamily: GAD domain-like
Family: GAD domain
Phyre2
| 14 |
|
PDB 2r7h chain A
Region: 88 - 106
Aligned: 18
Modelled: 19
Confidence: 6.8%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:putative d-alanine n-acetyltransferase of gnat family;
PDBTitle: crystal structure of a putative acetyltransferase of the gnat family2 (dde_3044) from desulfovibrio desulfuricans subsp. at 1.85 a3 resolution
Phyre2
| 15 |
|
PDB 2zw3 chain B
Region: 184 - 202
Aligned: 19
Modelled: 19
Confidence: 6.5%
Identity: 32%
PDB header:cell adhesion
Chain: B: PDB Molecule:gap junction beta-2 protein;
PDBTitle: structure of the connexin-26 gap junction channel at 3.52 angstrom resolution
Phyre2
| 16 |
|
PDB 2cnt chain D
Region: 88 - 106
Aligned: 18
Modelled: 19
Confidence: 6.4%
Identity: 28%
PDB header:transferase
Chain: D: PDB Molecule:modification of 30s ribosomal subunit protein s18;
PDBTitle: rimi - ribosomal s18 n-alpha-protein acetyltransferase in2 complex with coenzymea.
Phyre2
| 17 |
|
PDB 2psw chain A
Region: 88 - 106
Aligned: 18
Modelled: 19
Confidence: 5.9%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:n-acetyltransferase 13;
PDBTitle: human mak3 homolog in complex with coa
Phyre2
| 18 |
|
PDB 1puz chain A
Region: 4 - 42
Aligned: 39
Modelled: 39
Confidence: 5.7%
Identity: 10%
Fold: YgfY-like
Superfamily: YgfY-like
Family: YgfY-like
Phyre2
| 19 |
|
PDB 2ree chain B
Region: 88 - 108
Aligned: 20
Modelled: 21
Confidence: 5.4%
Identity: 30%
PDB header:transferase, lyase
Chain: B: PDB Molecule:cura;
PDBTitle: crystal structure of the loading gnatl domain of cura from lyngbya2 majuscula
Phyre2
| 20 |
|
PDB 1ev0 chain A
Region: 104 - 110
Aligned: 7
Modelled: 7
Confidence: 5.4%
Identity: 57%
Fold: Cell division protein MinE topological specificity domain
Superfamily: Cell division protein MinE topological specificity domain
Family: Cell division protein MinE topological specificity domain
Phyre2
| 21 |
|
| 22 |
|