| 1 |
|
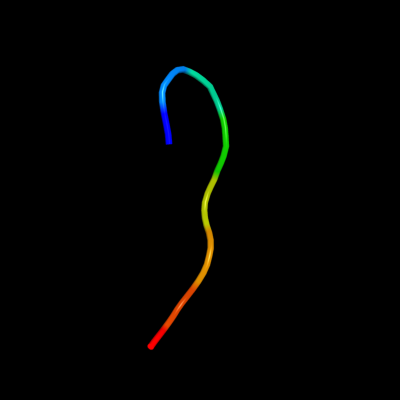
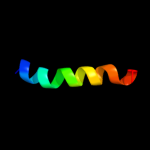
PDB 2jx5 chain A
Region: 44 - 51
Aligned: 8
Modelled: 8
Confidence: 9.8%
Identity: 38%
PDB header:ribosomal protein
Chain: A: PDB Molecule:glub(s27a);
PDBTitle: solution structure of the ubiquitin domain n-terminal to2 the s27a ribosomal subunit of giardia lamblia
Phyre2
| 2 |
|
PDB 2y69 chain Z
Region: 55 - 76
Aligned: 22
Modelled: 22
Confidence: 9.0%
Identity: 23%
PDB header:electron transport
Chain: Z: PDB Molecule:cytochrome c oxidase polypeptide 8h;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 3 |
|
PDB 1v54 chain M
Region: 55 - 76
Aligned: 22
Modelled: 22
Confidence: 8.8%
Identity: 23%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Family: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Phyre2
| 4 |
|
PDB 3nzz chain A
Region: 34 - 53
Aligned: 20
Modelled: 20
Confidence: 6.9%
Identity: 55%
PDB header:cell invasion
Chain: A: PDB Molecule:cell invasion protein sipd;
PDBTitle: crystal structure of the salmonella type iii secretion system tip2 protein sipd
Phyre2
| 5 |
|
PDB 1g2c chain N
Region: 40 - 54
Aligned: 15
Modelled: 15
Confidence: 6.1%
Identity: 47%
PDB header:viral protein
Chain: N: PDB Molecule:fusion protein (f);
PDBTitle: human respiratory syncytial virus fusion protein core
Phyre2
| 6 |
|
PDB 1sdi chain A
Region: 36 - 46
Aligned: 11
Modelled: 11
Confidence: 5.2%
Identity: 27%
Fold: YcfC-like
Superfamily: YcfC-like
Family: YcfC-like
Phyre2
| 7 |
|
PDB 2goh chain A
Region: 26 - 38
Aligned: 13
Modelled: 13
Confidence: 5.0%
Identity: 54%
PDB header:viral protein
Chain: A: PDB Molecule:vpu protein;
PDBTitle: three-dimensional structure of the trans-membrane domain of2 vpu from hiv-1 in aligned phospholipid bicelles
Phyre2
| 8 |
|
PDB 2gof chain A
Region: 26 - 38
Aligned: 13
Modelled: 13
Confidence: 5.0%
Identity: 54%
PDB header:viral protein
Chain: A: PDB Molecule:vpu protein;
PDBTitle: three-dimensional structure of the trans-membrane domain of2 vpu from hiv-1 in aligned phospholipid bicelles
Phyre2
| 9 |
|
PDB 1pje chain A
Region: 26 - 38
Aligned: 13
Modelled: 13
Confidence: 5.0%
Identity: 54%
PDB header:viral protein
Chain: A: PDB Molecule:vpu protein;
PDBTitle: structure of the channel-forming trans-membrane domain of2 virus protein "u"(vpu) from hiv-1
Phyre2
| 10 |
|
PDB 1pi7 chain A
Region: 26 - 38
Aligned: 13
Modelled: 13
Confidence: 5.0%
Identity: 54%
PDB header:viral protein
Chain: A: PDB Molecule:vpu protein;
PDBTitle: structure of the channel-forming trans-membrane domain of2 virus protein "u" (vpu) from hiv-1
Phyre2