| 1 |
|
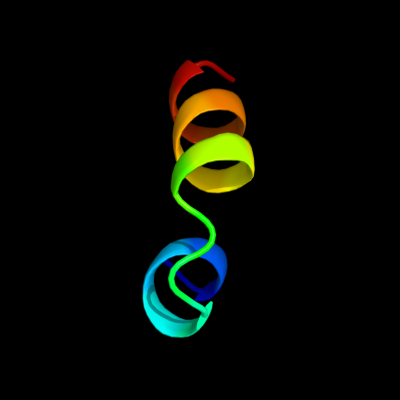
PDB 3iee chain A
Region: 23 - 40
Aligned: 18
Modelled: 18
Confidence: 13.3%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of hypothetical protein bf3319 from bacteroides2 fragilis (yp_212931.1) from bacteroides fragilis nctc 9343 at 1.70 a3 resolution
Phyre2
| 2 |
|
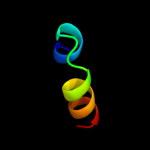
PDB 2axp chain A domain 1
Region: 31 - 47
Aligned: 17
Modelled: 17
Confidence: 8.7%
Identity: 18%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Nucleotide and nucleoside kinases
Phyre2
| 3 |
|
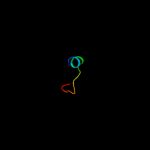
PDB 2f1f chain A domain 2
Region: 35 - 54
Aligned: 20
Modelled: 20
Confidence: 8.3%
Identity: 10%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: IlvH-like
Phyre2
| 4 |
|
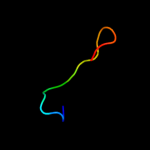
PDB 2fgc chain A domain 1
Region: 35 - 54
Aligned: 20
Modelled: 20
Confidence: 7.5%
Identity: 15%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: IlvH-like
Phyre2
| 5 |
|
PDB 3m92 chain B
Region: 25 - 37
Aligned: 13
Modelled: 13
Confidence: 7.5%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein ycin;
PDBTitle: the structure of ycin, an unchracterized protein from shigella2 flexneri.
Phyre2
| 6 |
|
PDB 2pc6 chain A domain 1
Region: 35 - 54
Aligned: 20
Modelled: 18
Confidence: 7.1%
Identity: 15%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: IlvH-like
Phyre2
| 7 |
|
PDB 1pjv chain A
Region: 27 - 41
Aligned: 15
Modelled: 15
Confidence: 7.1%
Identity: 33%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 8 |
|
PDB 1pu1 chain A
Region: 24 - 34
Aligned: 11
Modelled: 11
Confidence: 5.7%
Identity: 27%
Fold: Hypothetical protein MTH677
Superfamily: Hypothetical protein MTH677
Family: Hypothetical protein MTH677
Phyre2
| 9 |
|
PDB 1zq1 chain C domain 1
Region: 48 - 54
Aligned: 7
Modelled: 7
Confidence: 5.5%
Identity: 57%
Fold: GatB/YqeY motif
Superfamily: GatB/YqeY motif
Family: GatB/GatE C-terminal domain-like
Phyre2
| 10 |
|
PDB 2fxp chain A
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 5.4%
Identity: 27%
PDB header:viral protein
Chain: A: PDB Molecule:spike glycoprotein;
PDBTitle: solution structure of the sars-coronavirus hr2 domain
Phyre2