| 1 |
|
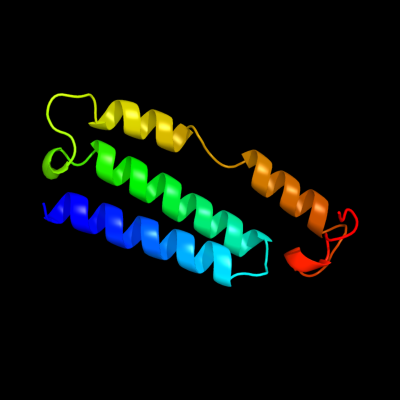
PDB 3rko chain K
Region: 125 - 216
Aligned: 91
Modelled: 92
Confidence: 98.6%
Identity: 23%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
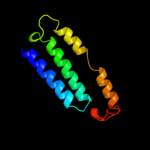
PDB 3rko chain N
Region: 24 - 216
Aligned: 183
Modelled: 193
Confidence: 20.7%
Identity: 14%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 3 |
|
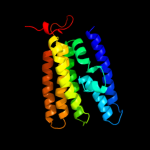
PDB 3rko chain M
Region: 88 - 214
Aligned: 125
Modelled: 127
Confidence: 20.2%
Identity: 12%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 4 |
|
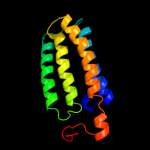
PDB 2jo1 chain A
Region: 115 - 146
Aligned: 32
Modelled: 32
Confidence: 15.2%
Identity: 28%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 5 |
|
PDB 2jp3 chain A
Region: 115 - 146
Aligned: 32
Modelled: 32
Confidence: 13.2%
Identity: 13%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 6 |
|
PDB 1v8c chain A domain 2
Region: 73 - 90
Aligned: 18
Modelled: 18
Confidence: 5.6%
Identity: 6%
Fold: TBP-like
Superfamily: MoaD-related protein, C-terminal domain
Family: MoaD-related protein, C-terminal domain
Phyre2