| 1 |
|
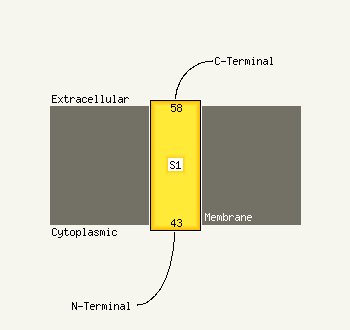
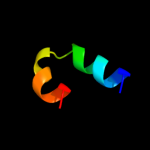
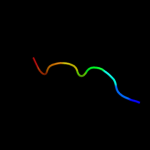
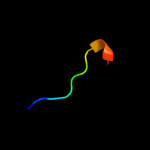
PDB 1gq2 chain A domain 2
Region: 26 - 53
Aligned: 28
Modelled: 28
Confidence: 27.2%
Identity: 32%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Malic enzyme N-domain
Phyre2
| 2 |
|
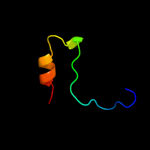
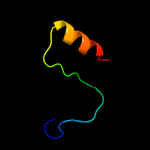
PDB 1pj3 chain A domain 2
Region: 26 - 53
Aligned: 28
Modelled: 28
Confidence: 21.8%
Identity: 25%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Malic enzyme N-domain
Phyre2
| 3 |
|
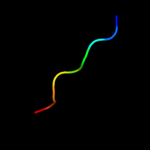
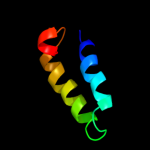
PDB 2aw5 chain A
Region: 26 - 53
Aligned: 28
Modelled: 28
Confidence: 14.4%
Identity: 29%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent malic enzyme;
PDBTitle: crystal structure of a human malic enzyme
Phyre2
| 4 |
|
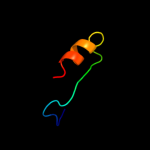
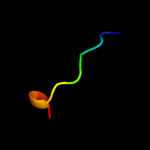
PDB 3ps9 chain A
Region: 42 - 61
Aligned: 20
Modelled: 20
Confidence: 13.1%
Identity: 50%
PDB header:transferase
Chain: A: PDB Molecule:trna 5-methylaminomethyl-2-thiouridine biosynthesis
PDBTitle: crystal structure of mnmc from e. coli
Phyre2
| 5 |
|
PDB 1gz3 chain B
Region: 26 - 53
Aligned: 28
Modelled: 28
Confidence: 11.9%
Identity: 25%
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent malic enzyme;
PDBTitle: molecular mechanism for the regulation of human mitochondrial2 nad(p)+-dependent malic enzyme by atp and fumarate
Phyre2
| 6 |
|
PDB 1dt0 chain A domain 1
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 11.3%
Identity: 56%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 7 |
|
PDB 1qr6 chain A
Region: 26 - 53
Aligned: 28
Modelled: 28
Confidence: 10.6%
Identity: 25%
PDB header:oxidoreductase
Chain: A: PDB Molecule:malic enzyme 2;
PDBTitle: human mitochondrial nad(p)-dependent malic enzyme
Phyre2
| 8 |
|
PDB 1y67 chain A domain 1
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 10.1%
Identity: 44%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 9 |
|
PDB 1my6 chain A domain 1
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 9.0%
Identity: 22%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 10 |
|
PDB 1zke chain A domain 1
Region: 19 - 41
Aligned: 23
Modelled: 23
Confidence: 8.8%
Identity: 30%
Fold: ROP-like
Superfamily: HP1531-like
Family: HP1531-like
Phyre2
| 11 |
|
PDB 1ix9 chain A domain 1
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 8.1%
Identity: 33%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 12 |
|
PDB 1reg chain X
Region: 33 - 54
Aligned: 22
Modelled: 22
Confidence: 8.0%
Identity: 27%
Fold: Ferredoxin-like
Superfamily: Translational regulator protein regA
Family: Translational regulator protein regA
Phyre2
| 13 |
|
PDB 1o0s chain A domain 2
Region: 22 - 53
Aligned: 32
Modelled: 32
Confidence: 7.8%
Identity: 16%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Malic enzyme N-domain
Phyre2
| 14 |
|
PDB 2j8b chain A domain 1
Region: 21 - 36
Aligned: 16
Modelled: 16
Confidence: 7.5%
Identity: 6%
Fold: Snake toxin-like
Superfamily: Snake toxin-like
Family: Extracellular domain of cell surface receptors
Phyre2
| 15 |
|
PDB 3rnv chain A
Region: 15 - 59
Aligned: 45
Modelled: 45
Confidence: 7.5%
Identity: 24%
PDB header:hydrolase
Chain: A: PDB Molecule:helper component proteinase;
PDBTitle: structure of the autocatalytic cysteine protease domain of potyvirus2 helper-component proteinase
Phyre2
| 16 |
|
PDB 2hdl chain A
Region: 32 - 39
Aligned: 8
Modelled: 8
Confidence: 7.1%
Identity: 50%
PDB header:cytokine
Chain: A: PDB Molecule:small inducible cytokine b14;
PDBTitle: solution structure of brak/cxcl14
Phyre2
| 17 |
|
PDB 2nyb chain A domain 1
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 7.1%
Identity: 44%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 18 |
|
PDB 3lrr chain B
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 7.1%
Identity: 27%
PDB header:hydrolase/rna
Chain: B: PDB Molecule:probable atp-dependent rna helicase ddx58;
PDBTitle: crystal structure of human rig-i ctd bound to a 12 bp au rich 5' ppp2 dsrna
Phyre2
| 19 |
|
PDB 2g6t chain A domain 1
Region: 33 - 48
Aligned: 16
Modelled: 16
Confidence: 6.8%
Identity: 25%
Fold: CAC2185-like
Superfamily: CAC2185-like
Family: CAC2185-like
Phyre2
| 20 |
|
PDB 3ga3 chain A
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 6.5%
Identity: 27%
PDB header:hydrolase
Chain: A: PDB Molecule:interferon-induced helicase c domain-containing
PDBTitle: crystal structure of the c-terminal domain of human mda5
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|