| 1 |
|
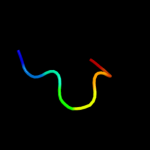
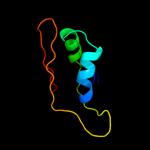
PDB 2hjj chain A domain 1
Region: 104 - 119
Aligned: 16
Modelled: 16
Confidence: 16.4%
Identity: 19%
Fold: dsRBD-like
Superfamily: YcfA/nrd intein domain
Family: YkfF-like
Phyre2
| 2 |
|
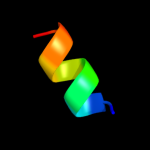
PDB 2hjj chain A
Region: 104 - 119
Aligned: 16
Modelled: 16
Confidence: 16.4%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ykff;
PDBTitle: solution nmr structure of protein ykff from escherichia coli.2 northeast structural genomics target er397.
Phyre2
| 3 |
|
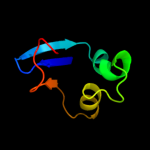
PDB 2vql chain D
Region: 129 - 136
Aligned: 8
Modelled: 8
Confidence: 11.1%
Identity: 75%
PDB header:membrane protein
Chain: D: PDB Molecule:uncharacterized protein cgl0972;
PDBTitle: crystal structure of porb from corynebacterium glutamicum (2 crystal form iii)
Phyre2
| 4 |
|
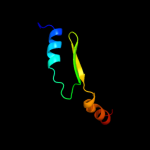
PDB 2rbd chain B
Region: 92 - 107
Aligned: 16
Modelled: 16
Confidence: 6.3%
Identity: 31%
PDB header:structural protein
Chain: B: PDB Molecule:bh2358 protein;
PDBTitle: crystal structure of a putative spore coat protein (bh2358) from2 bacillus halodurans c-125 at 1.54 a resolution
Phyre2
| 5 |
|
PDB 1af7 chain A
Region: 23 - 30
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 13%
PDB header:methyltransferase
Chain: A: PDB Molecule:chemotaxis receptor methyltransferase cher;
PDBTitle: cher from salmonella typhimurium
Phyre2
| 6 |
|
PDB 2ijr chain A domain 1
Region: 17 - 26
Aligned: 10
Modelled: 10
Confidence: 5.8%
Identity: 50%
Fold: Api92-like
Superfamily: Api92-like
Family: Api92-like
Phyre2
| 7 |
|
PDB 2c9w chain C domain 1
Region: 68 - 121
Aligned: 40
Modelled: 54
Confidence: 5.7%
Identity: 18%
Fold: POZ domain
Superfamily: POZ domain
Family: BTB/POZ domain
Phyre2
| 8 |
|
PDB 2ii8 chain F
Region: 49 - 75
Aligned: 27
Modelled: 27
Confidence: 5.5%
Identity: 19%
PDB header:signaling protein
Chain: F: PDB Molecule:anabaena sensory rhodopsin transducer protein;
PDBTitle: anabaena sensory rhodopsin transducer
Phyre2
| 9 |
|
PDB 1f7u chain A domain 3
Region: 78 - 130
Aligned: 53
Modelled: 53
Confidence: 5.4%
Identity: 9%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domain
Family: Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domain
Phyre2
| 10 |
|
PDB 2bjg chain B
Region: 7 - 71
Aligned: 65
Modelled: 65
Confidence: 5.4%
Identity: 14%
PDB header:hydrolase
Chain: B: PDB Molecule:choloylglycine hydrolase;
PDBTitle: crystal structure of conjugated bile acid hydrolase from2 clostridium perfringens in complex with reaction products3 taurine and deoxycholate
Phyre2
| 11 |
|
PDB 1kn7 chain A
Region: 23 - 40
Aligned: 18
Modelled: 18
Confidence: 5.2%
Identity: 44%
PDB header:membrane protein
Chain: A: PDB Molecule:voltage-gated potassium channel protein kv1.4;
PDBTitle: solution structure of the tandem inactivation domain2 (residues 1-75) of potassium channel rck4 (kv1.4)
Phyre2