| 1 |
|
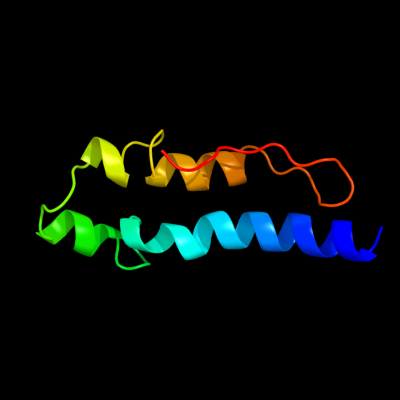
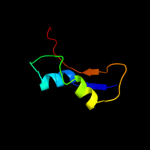
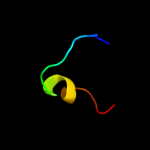
PDB 2kz6 chain A
Region: 61 - 132
Aligned: 61
Modelled: 72
Confidence: 98.3%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein cv0426 from chromobacterium violaceum,2 northeast structural genomics consortium (nesg) target cvt2
Phyre2
| 2 |
|
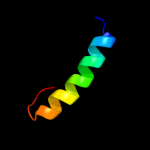
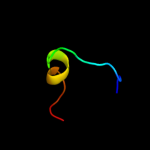
PDB 3o0r chain C
Region: 97 - 129
Aligned: 25
Modelled: 28
Confidence: 41.9%
Identity: 28%
PDB header:immune system/oxidoreductase
Chain: C: PDB Molecule:nitric oxide reductase subunit c;
PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
Phyre2
| 3 |
|
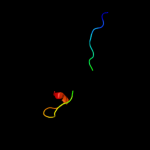
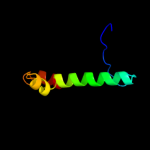
PDB 1fcq chain A
Region: 1 - 71
Aligned: 68
Modelled: 71
Confidence: 17.6%
Identity: 22%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Bee venom hyaluronidase
Phyre2
| 4 |
|
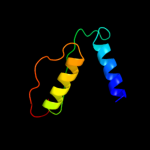
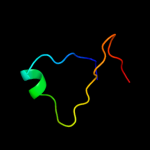
PDB 1d4b chain A
Region: 22 - 51
Aligned: 30
Modelled: 30
Confidence: 14.1%
Identity: 23%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: CAD domain
Phyre2
| 5 |
|
PDB 2atm chain A
Region: 1 - 54
Aligned: 51
Modelled: 54
Confidence: 10.9%
Identity: 27%
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronoglucosaminidase;
PDBTitle: crystal structure of the recombinant allergen ves v 2
Phyre2
| 6 |
|
PDB 1bgv chain A domain 2
Region: 97 - 121
Aligned: 25
Modelled: 25
Confidence: 8.9%
Identity: 8%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Aminoacid dehydrogenases
Phyre2
| 7 |
|
PDB 1twf chain C domain 1
Region: 5 - 25
Aligned: 21
Modelled: 21
Confidence: 8.7%
Identity: 24%
Fold: DCoH-like
Superfamily: RBP11-like subunits of RNA polymerase
Family: RNA polymerase alpha subunit dimerisation domain
Phyre2
| 8 |
|
PDB 2q22 chain A domain 1
Region: 69 - 130
Aligned: 58
Modelled: 62
Confidence: 8.3%
Identity: 16%
Fold: Ava3019-like
Superfamily: Ava3019-like
Family: Ava3019-like
Phyre2
| 9 |
|
PDB 1zak chain A domain 2
Region: 127 - 137
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 18%
Fold: Rubredoxin-like
Superfamily: Microbial and mitochondrial ADK, insert "zinc finger" domain
Family: Microbial and mitochondrial ADK, insert "zinc finger" domain
Phyre2
| 10 |
|
PDB 2ahm chain G
Region: 113 - 129
Aligned: 17
Modelled: 17
Confidence: 6.8%
Identity: 24%
PDB header:viral protein, replication
Chain: G: PDB Molecule:replicase polyprotein 1ab, heavy chain;
PDBTitle: crystal structure of sars-cov super complex of non-structural2 proteins: the hexadecamer
Phyre2
| 11 |
|
PDB 1ibx chain B
Region: 22 - 39
Aligned: 18
Modelled: 18
Confidence: 6.3%
Identity: 17%
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:chimera of igg binding protein g and dna
PDBTitle: nmr structure of dff40 and dff45 n-terminal domain complex
Phyre2
| 12 |
|
PDB 1ibx chain B
Region: 22 - 39
Aligned: 18
Modelled: 18
Confidence: 6.3%
Identity: 17%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: CAD domain
Phyre2
| 13 |
|
PDB 1x4t chain A domain 1
Region: 49 - 107
Aligned: 53
Modelled: 59
Confidence: 6.0%
Identity: 19%
Fold: Long alpha-hairpin
Superfamily: ISY1 domain-like
Family: ISY1 N-terminal domain-like
Phyre2
| 14 |
|
PDB 2eel chain A
Region: 22 - 52
Aligned: 31
Modelled: 31
Confidence: 5.5%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:cell death activator cide-a;
PDBTitle: solution structure of the cide-n domain of human cell death2 activator cide-a
Phyre2