| 1 |
|
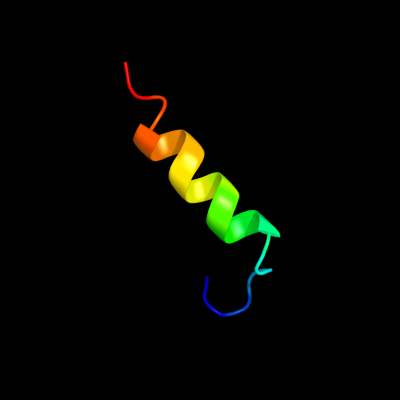
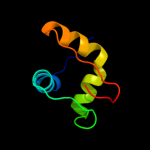
PDB 2k5c chain A
Region: 53 - 72
Aligned: 20
Modelled: 20
Confidence: 12.9%
Identity: 45%
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein pf0385;
PDBTitle: nmr structure for pf0385
Phyre2
| 2 |
|
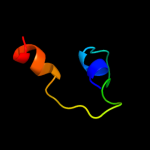
PDB 3otb chain B
Region: 34 - 88
Aligned: 55
Modelled: 55
Confidence: 10.7%
Identity: 24%
PDB header:transferase
Chain: B: PDB Molecule:trna(his) guanylyltransferase;
PDBTitle: crystal structure of human trnahis guanylyltransferase (thg1) - dgtp2 complex
Phyre2
| 3 |
|
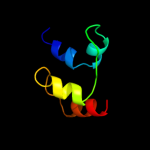
PDB 1gm5 chain A domain 1
Region: 96 - 106
Aligned: 11
Modelled: 11
Confidence: 9.1%
Identity: 45%
Fold: Four-helical up-and-down bundle
Superfamily: RecG, N-terminal domain
Family: RecG, N-terminal domain
Phyre2
| 4 |
|
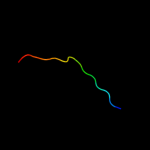
PDB 1oey chain J
Region: 76 - 95
Aligned: 20
Modelled: 20
Confidence: 7.6%
Identity: 20%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 5 |
|
PDB 1kdx chain B
Region: 38 - 45
Aligned: 8
Modelled: 8
Confidence: 7.3%
Identity: 63%
PDB header:transcription regulation complex
Chain: B: PDB Molecule:creb;
PDBTitle: kix domain of mouse cbp (creb binding protein) in complex2 with phosphorylated kinase inducible domain (pkid) of rat3 creb (cyclic amp response element binding protein), nmr 174 structures
Phyre2
| 6 |
|
PDB 3euk chain L
Region: 35 - 94
Aligned: 59
Modelled: 60
Confidence: 7.3%
Identity: 20%
PDB header:cell cycle
Chain: L: PDB Molecule:chromosome partition protein muke;
PDBTitle: crystal structure of muke-mukf(residues 292-443)-mukb(head2 domain)-atpgammas complex, asymmetric dimer
Phyre2
| 7 |
|
PDB 3ejh chain B
Region: 49 - 62
Aligned: 14
Modelled: 14
Confidence: 7.2%
Identity: 29%
PDB header:cell adhesion
Chain: B: PDB Molecule:fibronectin;
PDBTitle: crystal structure of the fibronectin 8-9fni domain pair in complex2 with a type-i collagen peptide
Phyre2
| 8 |
|
PDB 1x4q chain A
Region: 42 - 72
Aligned: 31
Modelled: 31
Confidence: 7.2%
Identity: 32%
PDB header:rna binding protein
Chain: A: PDB Molecule:u4/u6 small nuclear ribonucleoprotein prp3;
PDBTitle: solution structure of pwi domain in u4/u6 small nuclear2 ribonucleoprotein prp3(hprp3)
Phyre2
| 9 |
|
PDB 3chm chain A
Region: 59 - 108
Aligned: 42
Modelled: 50
Confidence: 6.5%
Identity: 24%
PDB header:plant protein
Chain: A: PDB Molecule:cop9 signalosome complex subunit 7;
PDBTitle: crystal structure of pci domain from a. thaliana cop9 signalosome2 subunit 7 (csn7)
Phyre2
| 10 |
|
PDB 2kmc chain A
Region: 80 - 90
Aligned: 11
Modelled: 11
Confidence: 6.3%
Identity: 45%
PDB header:cell adhesion
Chain: A: PDB Molecule:fermitin family homolog 1;
PDBTitle: solution structure of the n-terminal domain of kindlin-1
Phyre2
| 11 |
|
PDB 3lr6 chain A
Region: 36 - 70
Aligned: 35
Modelled: 35
Confidence: 5.8%
Identity: 20%
PDB header:structural protein
Chain: A: PDB Molecule:major ampullate spidroin 1;
PDBTitle: self-assembly of spider silk proteins is controlled by a ph-sensitive2 relay
Phyre2
| 12 |
|
PDB 3n7m chain A
Region: 82 - 96
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 33%
PDB header:toxin
Chain: A: PDB Molecule:neurotoxin;
PDBTitle: crystal structure of w1252a mutant of hcr d/c vpi 5995
Phyre2