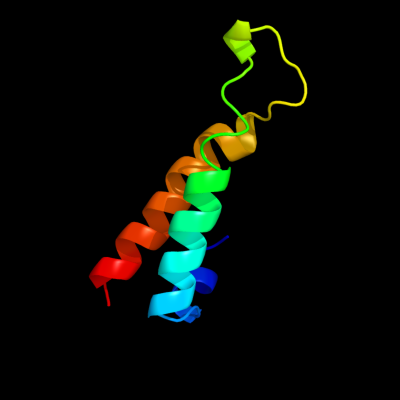
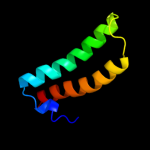
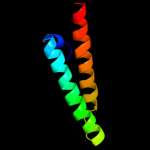
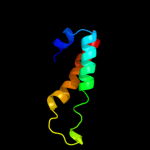
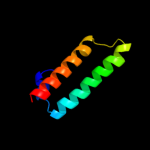
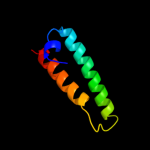
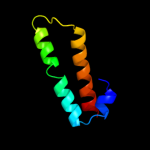
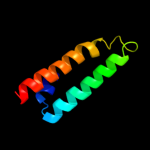
1 d1r73a_
99.9
44
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)2 d2zjrv1
99.9
38
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)3 c2j375_
99.9
32
PDB header: ribosomeChain: 5: PDB Molecule: ribosomal protein l35;PDBTitle: model of mammalian srp bound to 80s rncs
4 d2gycw1
99.9
100
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)5 d1vqov1
99.9
33
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)6 c2zkrv_
99.9
36
PDB header: ribosomal protein/rnaChain: V: PDB Molecule: rna expansion segment es9 part2;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
7 c3bboZ_
99.9
32
PDB header: ribosomeChain: Z: PDB Molecule: ribosomal protein l29;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
8 d2hgq11
99.9
39
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)9 c3jywX_
99.9
38
PDB header: ribosomeChain: X: PDB Molecule: 60s ribosomal protein l35;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
10 c1s1iX_
99.8
40
PDB header: ribosomeChain: X: PDB Molecule: 60s ribosomal protein l35;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
11 c4a1eU_
99.7
29
PDB header: ribosomeChain: U: PDB Molecule: rpl35;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
12 d2j0121
99.3
39
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)13 c2a8vA_
26.0
38
PDB header: protein/rnaChain: A: PDB Molecule: rna binding domain of rho transcriptionPDBTitle: rho transcription termination factor/rna complex
14 c2hv8D_
21.0
26
PDB header: protein transportChain: D: PDB Molecule: rab11 family-interacting protein 3;PDBTitle: crystal structure of gtp-bound rab11 in complex with fip3
15 c2o1sC_
18.0
26
PDB header: transferaseChain: C: PDB Molecule: 1-deoxy-d-xylulose-5-phosphate synthase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate synthase (dxs) from2 escherichia coli
16 d1gjja2
14.9
25
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain17 d1xwya1
13.9
11
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: TatD Mg-dependent DNase-like18 c2o1xA_
12.9
27
PDB header: transferaseChain: A: PDB Molecule: 1-deoxy-d-xylulose-5-phosphate synthase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate synthase (dxs) from2 deinococcus radiodurans
19 c2xusA_
11.7
19
PDB header: protein bindingChain: A: PDB Molecule: breast cancer metastasis-suppressor 1;PDBTitle: crystal structure of the brms1 n-terminal region
20 c1sy9B_
11.2
42
PDB header: calcium-binding proteinChain: B: PDB Molecule: cyclic-nucleotide-gated olfactory channel;PDBTitle: structure of calmodulin complexed with a fragment of the2 olfactory cng channel
21 c1envA_
not modelled
11.0
26
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 envelope protein chimera consisting of a fragment ofPDBTitle: atomic structure of the ectodomain from hiv-1 gp41
22 c1gcmC_
not modelled
10.9
25
PDB header: transcription regulationChain: C: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
23 d2odgc1
not modelled
10.8
31
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain24 c2rg8A_
not modelled
9.4
16
PDB header: apoptosis, translationChain: A: PDB Molecule: programmed cell death protein 4;PDBTitle: crystal structure of programmed for cell death 4 middle ma32 domain
25 d1jeia_
not modelled
8.5
36
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain26 c1kd8E_
not modelled
8.3
22
PDB header: de novo proteinChain: E: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
27 c1j1eB_
not modelled
6.9
17
PDB header: contractile proteinChain: B: PDB Molecule: troponin t;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
28 d1r8ja1
not modelled
6.8
15
Fold: KaiA/RbsU domainSuperfamily: KaiA/RbsU domainFamily: Circadian clock protein KaiA, C-terminal domain29 c1ce9C_
not modelled
6.6
35
PDB header: helix cappingChain: C: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
30 c1ce9A_
not modelled
6.6
35
PDB header: helix cappingChain: A: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
31 c1ce9B_
not modelled
6.6
35
PDB header: helix cappingChain: B: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
32 c1ce9D_
not modelled
6.6
35
PDB header: helix cappingChain: D: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
33 d1dd3a1
not modelled
6.2
17
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain34 c2wq2A_
not modelled
6.2
13
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with three ixxntxx motifs2 coordinating iodide
35 c3czqA_
not modelled
6.1
19
PDB header: transferaseChain: A: PDB Molecule: putative polyphosphate kinase 2;PDBTitle: crystal structure of putative polyphosphate kinase 2 from2 sinorhizobium meliloti
36 d1ov9a_
not modelled
6.1
35
Fold: H-NS histone-like proteinsSuperfamily: H-NS histone-like proteinsFamily: H-NS histone-like proteins37 d1sv1a_
not modelled
6.0
15
Fold: KaiA/RbsU domainSuperfamily: KaiA/RbsU domainFamily: Circadian clock protein KaiA, C-terminal domain38 d2a1ja1
not modelled
6.0
23
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like39 c2wq0A_
not modelled
5.9
13
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with three ixxntxx motifs2 coordinating chloride
40 d1hf8a1
not modelled
5.8
17
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: Phosphoinositide-binding clathrin adaptor, domain 241 d1h1js_
not modelled
5.8
19
Fold: LEM/SAP HeH motifSuperfamily: SAP domainFamily: SAP domain42 d2oa5a1
not modelled
5.7
32
Fold: BLRF2-likeSuperfamily: BLRF2-likeFamily: BLRF2-like43 c2kztA_
not modelled
5.7
16
PDB header: apoptosisChain: A: PDB Molecule: programmed cell death protein 4;PDBTitle: structure of the tandem ma-3 region of pdcd4
44 d1zrja1
not modelled
5.6
38
Fold: LEM/SAP HeH motifSuperfamily: SAP domainFamily: SAP domain45 d1hx8a1
not modelled
5.3
30
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: Phosphoinositide-binding clathrin adaptor, domain 246 c2outA_
not modelled
5.1
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mu-like prophage flumu protein gp35, proteinPDBTitle: solution structure of hi1506, a novel two domain protein2 from haemophilus influenzae
47 c1gcmB_
not modelled
5.1
26
PDB header: transcription regulationChain: B: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
48 c3layF_
not modelled
5.1
14
PDB header: metal binding proteinChain: F: PDB Molecule: zinc resistance-associated protein;PDBTitle: alpha-helical barrel formed by the decamer of the zinc resistance-2 associated protein (stm4172) from salmonella enterica subsp. enterica3 serovar typhimurium str. lt2
49 c2jo8B_
not modelled
5.1
35
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase 4;PDBTitle: solution structure of c-terminal domain of human mammalian2 sterile 20-like kinase 1 (mst1)