| 1 |
|
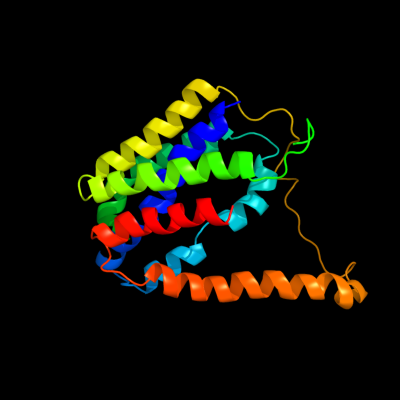
PDB 2gfp chain A
Region: 5 - 219
Aligned: 215
Modelled: 215
Confidence: 60.0%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 2 |
|
PDB 2knc chain A
Region: 129 - 162
Aligned: 34
Modelled: 34
Confidence: 13.4%
Identity: 12%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 3 |
|
PDB 2oar chain A domain 1
Region: 232 - 252
Aligned: 21
Modelled: 21
Confidence: 11.7%
Identity: 29%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 4 |
|
PDB 3lr6 chain A
Region: 19 - 57
Aligned: 39
Modelled: 39
Confidence: 10.0%
Identity: 18%
PDB header:structural protein
Chain: A: PDB Molecule:major ampullate spidroin 1;
PDBTitle: self-assembly of spider silk proteins is controlled by a ph-sensitive2 relay
Phyre2
| 5 |
|
PDB 3rko chain M
Region: 126 - 204
Aligned: 78
Modelled: 79
Confidence: 9.8%
Identity: 12%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 6 |
|
PDB 1wgl chain A
Region: 223 - 246
Aligned: 24
Modelled: 24
Confidence: 7.3%
Identity: 17%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: CUE domain
Phyre2
| 7 |
|
PDB 3mk7 chain F
Region: 124 - 193
Aligned: 70
Modelled: 70
Confidence: 5.9%
Identity: 16%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 8 |
|
PDB 1dwk chain A domain 1
Region: 27 - 62
Aligned: 36
Modelled: 36
Confidence: 5.6%
Identity: 8%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: Cyanase N-terminal domain
Phyre2