| 1 |
|
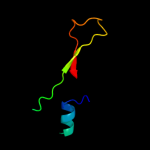
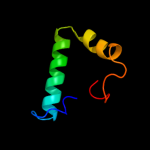
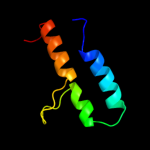
PDB 1kbh chain B
Region: 13 - 54
Aligned: 42
Modelled: 42
Confidence: 63.9%
Identity: 24%
Fold: Nuclear receptor coactivator interlocking domain
Superfamily: Nuclear receptor coactivator interlocking domain
Family: Nuclear receptor coactivator interlocking domain
Phyre2
| 2 |
|
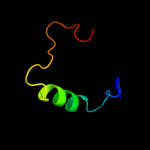
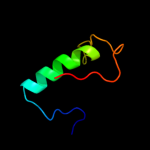
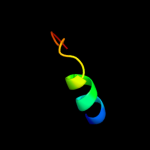
PDB 1a9x chain A domain 1
Region: 155 - 192
Aligned: 38
Modelled: 38
Confidence: 37.1%
Identity: 24%
Fold: Carbamoyl phosphate synthetase, large subunit connection domain
Superfamily: Carbamoyl phosphate synthetase, large subunit connection domain
Family: Carbamoyl phosphate synthetase, large subunit connection domain
Phyre2
| 3 |
|
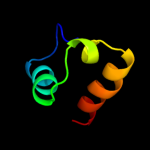
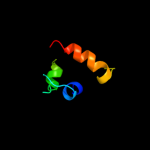
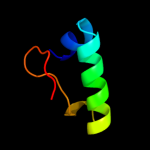
PDB 3c7c chain B
Region: 99 - 214
Aligned: 116
Modelled: 116
Confidence: 25.5%
Identity: 10%
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
Phyre2
| 4 |
|
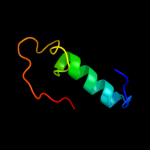
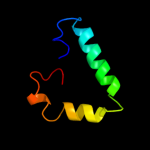
PDB 1m6v chain E
Region: 154 - 192
Aligned: 39
Modelled: 39
Confidence: 24.3%
Identity: 23%
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
Phyre2
| 5 |
|
PDB 2xu8 chain B
Region: 98 - 134
Aligned: 36
Modelled: 37
Confidence: 19.3%
Identity: 19%
PDB header:structural genomics
Chain: B: PDB Molecule:pa1645;
PDBTitle: structure of pa1645
Phyre2
| 6 |
|
PDB 1ej5 chain A
Region: 14 - 52
Aligned: 39
Modelled: 39
Confidence: 19.2%
Identity: 18%
Fold: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Superfamily: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Family: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Phyre2
| 7 |
|
PDB 1ega chain A domain 2
Region: 34 - 74
Aligned: 35
Modelled: 41
Confidence: 12.2%
Identity: 31%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Prokaryotic type KH domain (KH-domain type II)
Family: Prokaryotic type KH domain (KH-domain type II)
Phyre2
| 8 |
|
PDB 1yvr chain A domain 1
Region: 97 - 146
Aligned: 50
Modelled: 50
Confidence: 10.0%
Identity: 14%
Fold: alpha-alpha superhelix
Superfamily: TROVE domain-like
Family: TROVE domain-like
Phyre2
| 9 |
|
PDB 1ega chain B
Region: 31 - 74
Aligned: 38
Modelled: 44
Confidence: 9.1%
Identity: 29%
PDB header:hydrolase
Chain: B: PDB Molecule:protein (gtp-binding protein era);
PDBTitle: crystal structure of a widely conserved gtpase era
Phyre2
| 10 |
|
PDB 1rh6 chain A
Region: 135 - 146
Aligned: 12
Modelled: 12
Confidence: 7.9%
Identity: 33%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Excisionase-like
Phyre2
| 11 |
|
PDB 1p9q chain C domain 1
Region: 129 - 171
Aligned: 42
Modelled: 42
Confidence: 7.8%
Identity: 21%
Fold: RuvA C-terminal domain-like
Superfamily: Hypothetical protein AF0491, middle domain
Family: Hypothetical protein AF0491, middle domain
Phyre2
| 12 |
|
PDB 1pm6 chain A
Region: 135 - 146
Aligned: 12
Modelled: 12
Confidence: 7.1%
Identity: 33%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Excisionase-like
Phyre2
| 13 |
|
PDB 1yvr chain A
Region: 97 - 146
Aligned: 50
Modelled: 50
Confidence: 6.9%
Identity: 18%
PDB header:rna binding protein
Chain: A: PDB Molecule:60-kda ss-a/ro ribonucleoprotein;
PDBTitle: ro autoantigen
Phyre2
| 14 |
|
PDB 1z0j chain B domain 1
Region: 37 - 91
Aligned: 47
Modelled: 55
Confidence: 6.8%
Identity: 23%
Fold: Long alpha-hairpin
Superfamily: Rabenosyn-5 Rab-binding domain-like
Family: Rabenosyn-5 Rab-binding domain-like
Phyre2
| 15 |
|
PDB 1sse chain B
Region: 161 - 177
Aligned: 17
Modelled: 17
Confidence: 6.7%
Identity: 24%
PDB header:transcription activator
Chain: B: PDB Molecule:ap-1 like transcription factor yap1;
PDBTitle: solution structure of the oxidized form of the yap1 redox2 domain
Phyre2
| 16 |
|
PDB 1gw4 chain A
Region: 67 - 87
Aligned: 21
Modelled: 21
Confidence: 6.7%
Identity: 38%
PDB header:high density lipoproteins
Chain: A: PDB Molecule:apoa-i;
PDBTitle: the helix-hinge-helix structural motif in human2 apolipoprotein a-i determined by nmr spectroscopy, 13 structure
Phyre2
| 17 |
|
PDB 1zoq chain C domain 1
Region: 43 - 54
Aligned: 12
Modelled: 12
Confidence: 6.1%
Identity: 42%
Fold: Nuclear receptor coactivator interlocking domain
Superfamily: Nuclear receptor coactivator interlocking domain
Family: Nuclear receptor coactivator interlocking domain
Phyre2
| 18 |
|
PDB 1oj4 chain A domain 2
Region: 65 - 103
Aligned: 35
Modelled: 39
Confidence: 6.0%
Identity: 14%
Fold: Ferredoxin-like
Superfamily: GHMP Kinase, C-terminal domain
Family: 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase IspE
Phyre2
| 19 |
|
PDB 1wf3 chain A domain 2
Region: 36 - 52
Aligned: 17
Modelled: 17
Confidence: 6.0%
Identity: 24%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Prokaryotic type KH domain (KH-domain type II)
Family: Prokaryotic type KH domain (KH-domain type II)
Phyre2
| 20 |
|
PDB 1g47 chain A domain 1
Region: 171 - 188
Aligned: 18
Modelled: 18
Confidence: 5.9%
Identity: 22%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 21 |
|