| 1 |
|
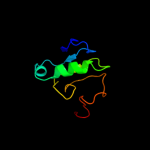
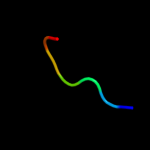
PDB 1uc2 chain A
Region: 3 - 83
Aligned: 81
Modelled: 81
Confidence: 99.9%
Identity: 26%
Fold: Hypothetical protein PH1602
Superfamily: Hypothetical protein PH1602
Family: Hypothetical protein PH1602
Phyre2
| 2 |
|
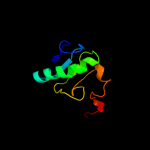
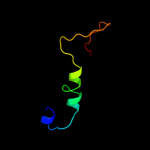
PDB 2epg chain B
Region: 3 - 83
Aligned: 79
Modelled: 81
Confidence: 99.9%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ttha1785;
PDBTitle: crystal structure of ttha1785
Phyre2
| 3 |
|
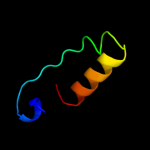
PDB 1wkq chain A
Region: 52 - 71
Aligned: 20
Modelled: 20
Confidence: 24.4%
Identity: 30%
Fold: Cytidine deaminase-like
Superfamily: Cytidine deaminase-like
Family: Deoxycytidylate deaminase-like
Phyre2
| 4 |
|
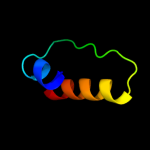
PDB 1w96 chain A domain 2
Region: 52 - 58
Aligned: 7
Modelled: 7
Confidence: 16.1%
Identity: 29%
Fold: PreATP-grasp domain
Superfamily: PreATP-grasp domain
Family: BC N-terminal domain-like
Phyre2
| 5 |
|
PDB 1ulz chain A domain 2
Region: 52 - 58
Aligned: 7
Modelled: 7
Confidence: 15.1%
Identity: 57%
Fold: PreATP-grasp domain
Superfamily: PreATP-grasp domain
Family: BC N-terminal domain-like
Phyre2
| 6 |
|
PDB 2j9g chain A domain 2
Region: 52 - 58
Aligned: 7
Modelled: 7
Confidence: 14.7%
Identity: 57%
Fold: PreATP-grasp domain
Superfamily: PreATP-grasp domain
Family: BC N-terminal domain-like
Phyre2
| 7 |
|
PDB 1el6 chain A
Region: 31 - 76
Aligned: 44
Modelled: 46
Confidence: 11.1%
Identity: 20%
Fold: Baseplate structural protein gp11
Superfamily: Baseplate structural protein gp11
Family: Baseplate structural protein gp11
Phyre2
| 8 |
|
PDB 1j8y chain F domain 2
Region: 8 - 43
Aligned: 34
Modelled: 36
Confidence: 10.1%
Identity: 15%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Nitrogenase iron protein-like
Phyre2
| 9 |
|
PDB 1jql chain B
Region: 4 - 38
Aligned: 35
Modelled: 35
Confidence: 6.4%
Identity: 9%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Extended AAA-ATPase domain
Phyre2
| 10 |
|
PDB 2zkq chain M
Region: 2 - 50
Aligned: 49
Modelled: 49
Confidence: 6.3%
Identity: 8%
PDB header:ribosomal protein/rna
Chain: M: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
Phyre2
| 11 |
|
PDB 2hxv chain A domain 2
Region: 57 - 71
Aligned: 15
Modelled: 15
Confidence: 6.3%
Identity: 33%
Fold: Cytidine deaminase-like
Superfamily: Cytidine deaminase-like
Family: Deoxycytidylate deaminase-like
Phyre2
| 12 |
|
PDB 2b7f chain D
Region: 4 - 41
Aligned: 38
Modelled: 38
Confidence: 5.4%
Identity: 21%
PDB header:hydrolase/hydrolase inhibitor
Chain: D: PDB Molecule:htlv protease;
PDBTitle: crystal structure of human t-cell leukemia virus protease, a novel2 target for anti-cancer design
Phyre2
| 13 |
|
PDB 1wwr chain A domain 1
Region: 57 - 71
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 27%
Fold: Cytidine deaminase-like
Superfamily: Cytidine deaminase-like
Family: Deoxycytidylate deaminase-like
Phyre2